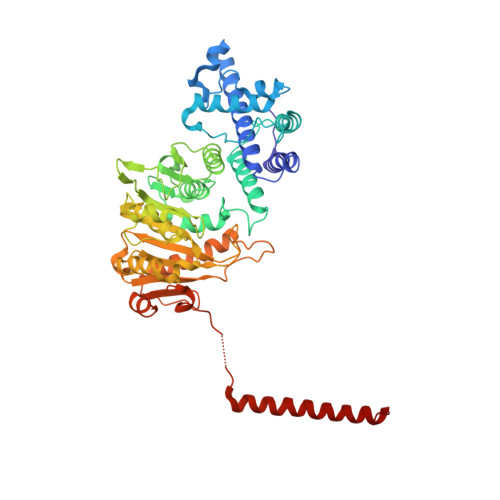
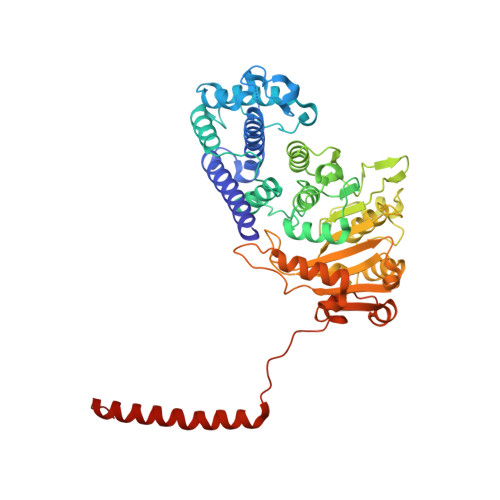
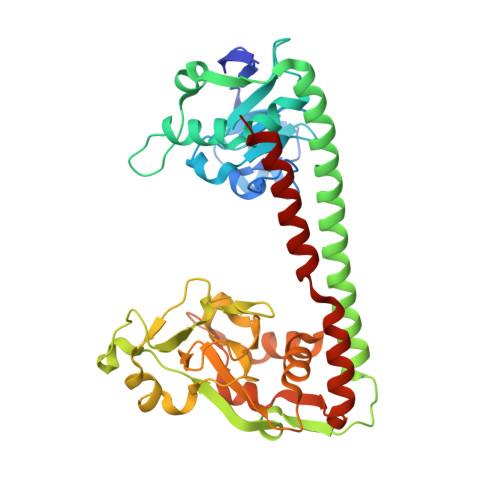
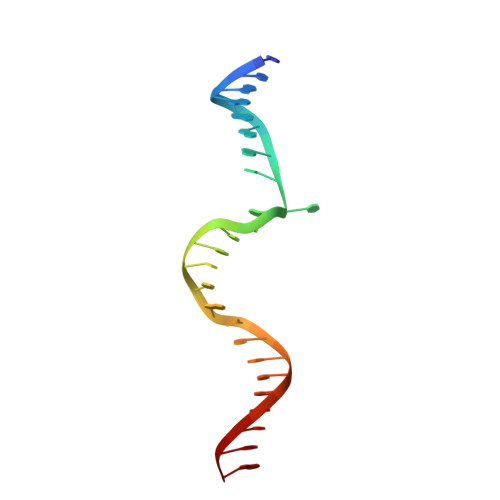
Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Zhu, J., Gao, Y., Wang, Y., Zhan, Q., Feng, H., Luo, X., Li, P., Liu, S., Hou, H., Gao, P.(2022) Nat Commun 13: 6391-6391
- PubMed: 36302770
- DOI: https://doi.org/10.1038/s41467-022-34085-z
- Primary Citation of Related Structures:
7VRU, 7VS4 - PubMed Abstract:
Type I restriction-modification systems help establish the prokaryotic DNA methylation landscape and provide protection against invasive DNA. In addition to classical m6A modifications, non-canonical type I enzymes catalyze both m6A and m4C using alternative DNA-modification subunits M1 and M2. Here, we report the crystal structures of the non-canonical PacII_M1M2S methyltransferase bound to target DNA and reaction product S-adenosylhomocysteine in a closed clamp-like conformation. Target DNA binds tightly within the central tunnel of the M1M2S complex and forms extensive contacts with all three protein subunits. Unexpectedly, while the target cytosine properly inserts into M2's pocket, the target adenine (either unmethylated or methylated) is anchored outside M1's pocket. A unique asymmetric catalysis is established where PacII_M1M2S has precisely coordinated the relative conformations of different subunits and evolved specific amino acids within M2/M1. This work provides insights into mechanisms of m6A/m4C catalysis and guidance for designing tools based on type I restriction-modification enzymes.
- CAS Key Laboratory of Infection and Immunity, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.
Organizational Affiliation: