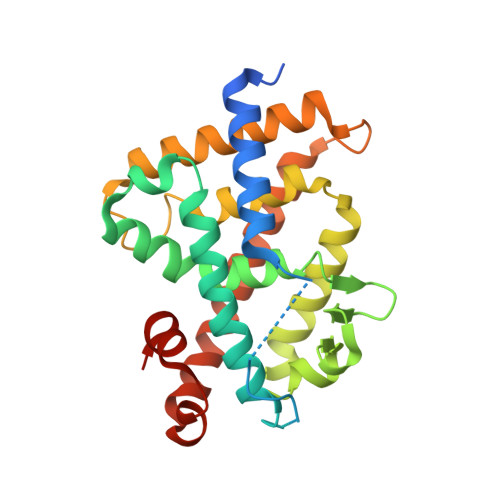
Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Yoshihara, A., Kawasaki, H., Masuno, H., Takada, K., Numoto, N., Ito, N., Hirata, N., Kanda, Y., Ishizawa, M., Makishima, M., Kagechika, H., Tanatani, A.(2022) Biomolecules 12
- PubMed: 35053278
- DOI: https://doi.org/10.3390/biom12010130
- Primary Citation of Related Structures:
7VQP - PubMed Abstract:
1α,25-Dihydroxyvitamin D 3 [1α,25(OH) 2 D 3 , 1 ] is an active form of vitamin D 3 and regulates various biological phenomena, including calcium and phosphate homeostasis, bone metabolism, and immune response via binding to and activation of vitamin D receptor (VDR). Lithocholic acid (LCA, 2 ) was identified as a second endogenous agonist of VDR, though its potency is very low. However, the lithocholic acid derivative 3 ( Dcha-20 ) is a more potent agonist than 1α,25(OH) 2 D 3 , ( 1 ), and its carboxyl group has similar interactions to the 1,3-dihydroxyl groups of 1 with amino acid residues in the VDR ligand-binding pocket. Here, we designed and synthesized amide derivatives of 3 in order to clarify the role of the carboxyl group. The synthesized amide derivatives showed HL-60 cell differentiation-inducing activity with potency that depended upon the substituent on the amide nitrogen atom. Among them, the N -cyanoamide 6 is more active than either 1 or 3 .
- Department of Chemistry, Faculty of Science, Ochanomizu University, 2-1-1 Otsuka, Bunkyo-ku, Tokyo 112-8610, Japan.
Organizational Affiliation: