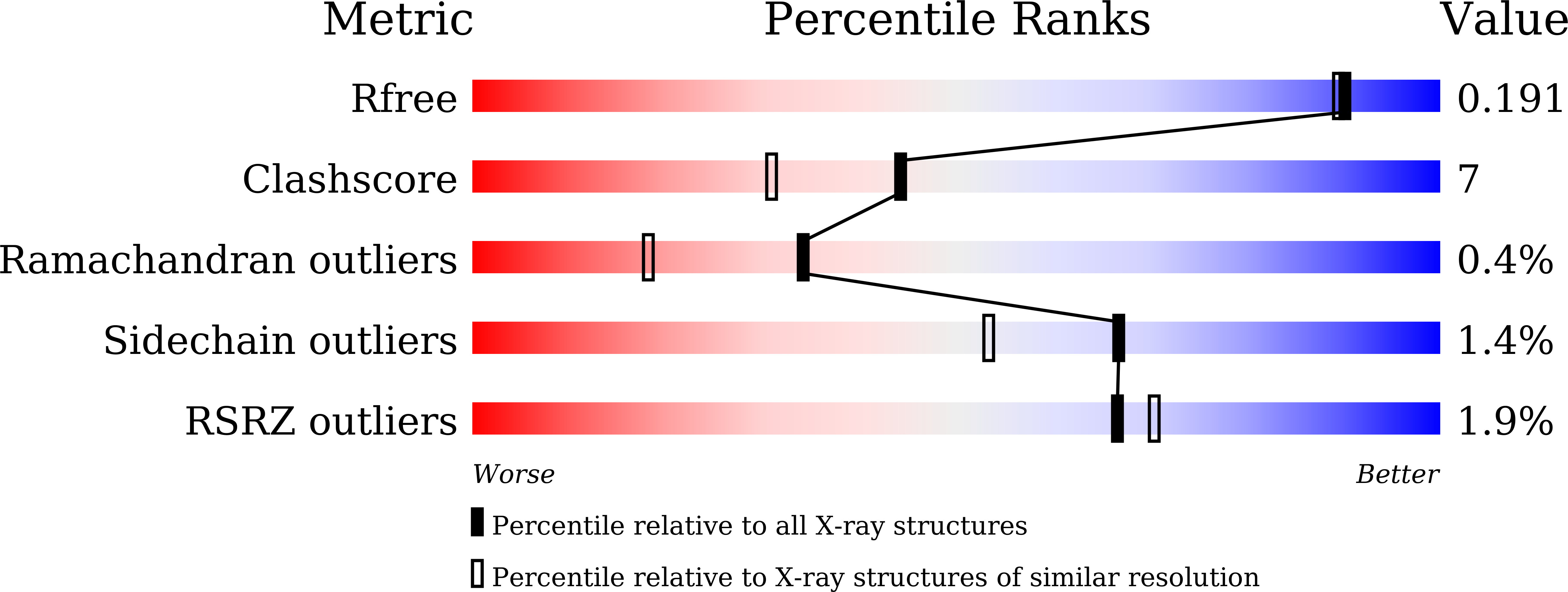
Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Meyer Cifuentes, I.E., Wu, P., Zhao, Y., Liu, W., Neumann-Schaal, M., Pfaff, L., Barys, J., Li, Z., Gao, J., Han, X., Bornscheuer, U.T., Wei, R., Ozturk, B.(2022) Front Bioeng Biotechnol 10: 930140-930140
- PubMed: 35935485
- DOI: https://doi.org/10.3389/fbioe.2022.930140
- Primary Citation of Related Structures:
7VMD, 7VPA - PubMed Abstract:
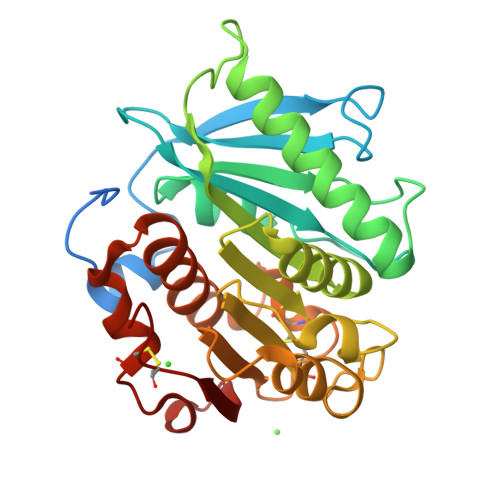
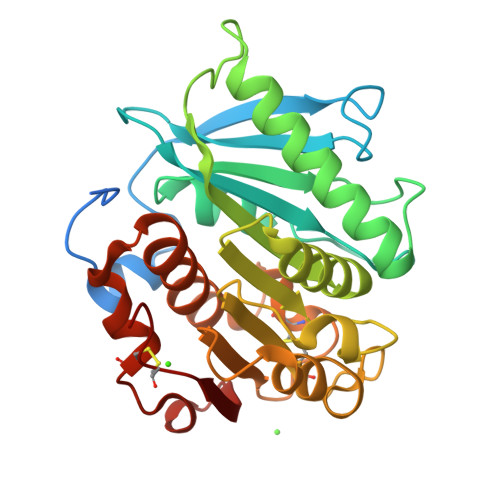
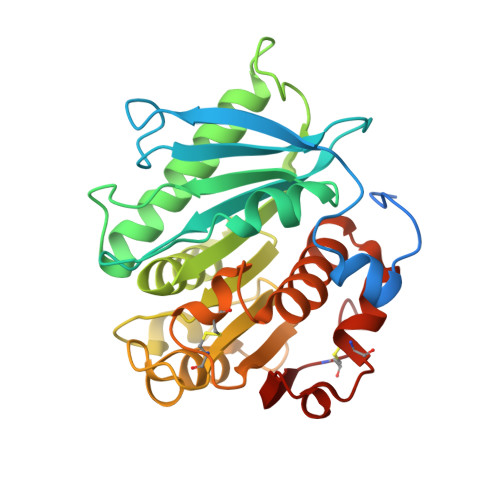
Polybutylene adipate terephthalate (PBAT) is a biodegradable alternative to polyethylene and can be broadly used in various applications. These polymers can be degraded by hydrolases of terrestrial and aquatic origin. In a previous study, we identified tandem PETase-like hydrolases (Ples) from the marine microbial consortium I1 that were highly expressed when a PBAT blend was supplied as the only carbon source. In this study, the tandem Ples, Ple628 and Ple629, were recombinantly expressed and characterized. Both enzymes are mesophilic and active on a wide range of oligomers. The activities of the Ples differed greatly when model substrates, PBAT-modified polymers or PET nanoparticles were supplied. Ple629 was always more active than Ple628. Crystal structures of Ple628 and Ple629 revealed a structural similarity to other PETases and can be classified as member of the PETases IIa subclass, α/β hydrolase superfamily. Our results show that the predicted functions of Ple628 and Ple629 agree with the bioinformatic predictions, and these enzymes play a significant role in the plastic degradation by the consortium.
Organizational Affiliation:
Junior Research Group Microbial Biotechnology, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.