Structural studies of a novel auxiliary-domain-containing phenylalanine hydroxylase from Bacillus cereus ATCC 14579.
Park, J., Hong, J., Seok, J., Hong, H., Seo, H., Kim, K.J.(2022) Acta Crystallogr D Struct Biol 78: 586-598
- PubMed: 35503207
- DOI: https://doi.org/10.1107/S2059798322002674
- Primary Citation of Related Structures:
7VGM - PubMed Abstract:
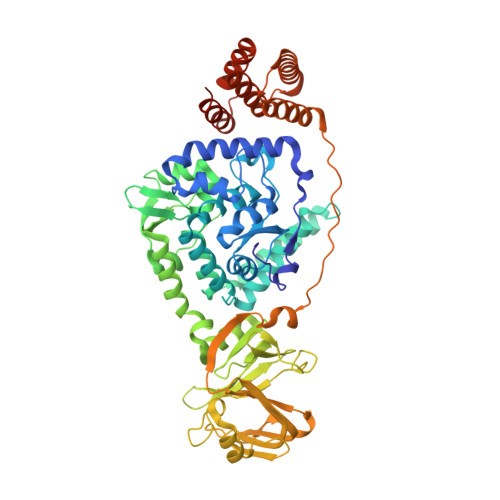
Phenylalanine hydroxylase (PAH), which belongs to the aromatic amino-acid hydroxylase family, is involved in protein synthesis and pyomelanine production through the hydroxylation of phenylalanine to tyrosine. In this study, the crystal structure of PAH from Bacillus cereus ATCC 14579 (BcPAH) with an additional 280 amino acids in the C-terminal region was determined. The structure of BcPAH consists of three distinct domains: a core domain with two additional inserted α-helices and two novel auxiliary domains: BcPAH-AD1 and BcPAH-AD2. Structural homologues of BcPAH-AD1 and BcPAH-AD2 are known to be involved in mRNA regulation and protein-protein interactions, and thus it was speculated that BcPAH might utilize the auxiliary domains for interaction with its partner proteins. Furthermore, phylogenetic tree analysis revealed that the three-domain PAHs, including BcPAH, are completely distinctive from both conventional prokaryotic PAHs and eukaryotic PAHs. Finally, biochemical studies of BcPAH showed that BcPAH-AD1 might be important for the structural integrity of the enzyme and that BcPAH-AD2 is related to enzyme stability and/or activity. Investigations into the intracellular functions of the two auxiliary domains and the relationship between these functions and the activity of PAH are required.
- School of Life Sciences, BK21 FOUR KNU Creative BioResearch Group, Kyungpook National University, Daegu 41566, Republic of Korea.
Organizational Affiliation: