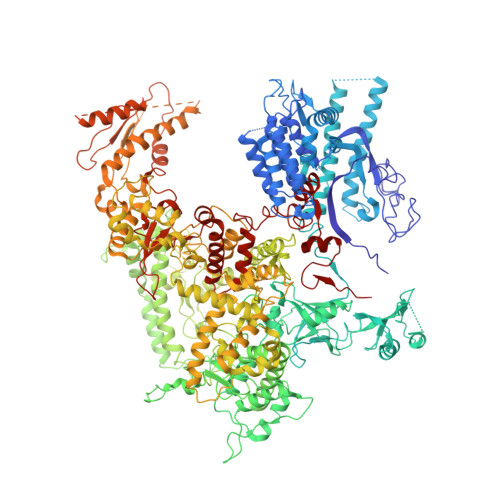
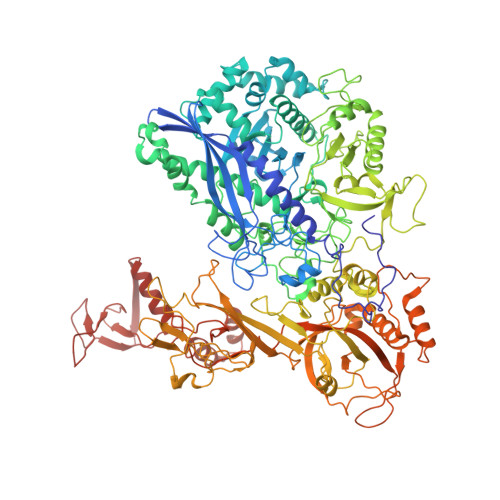
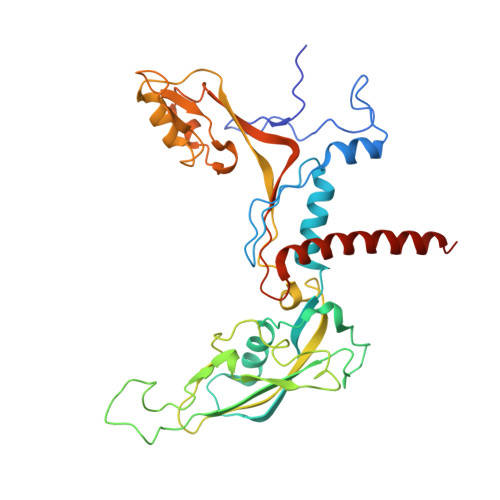
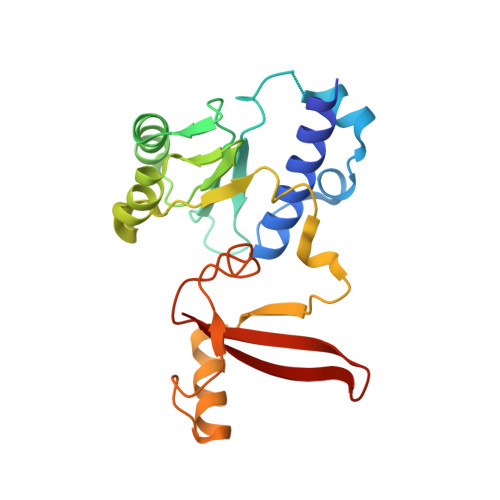
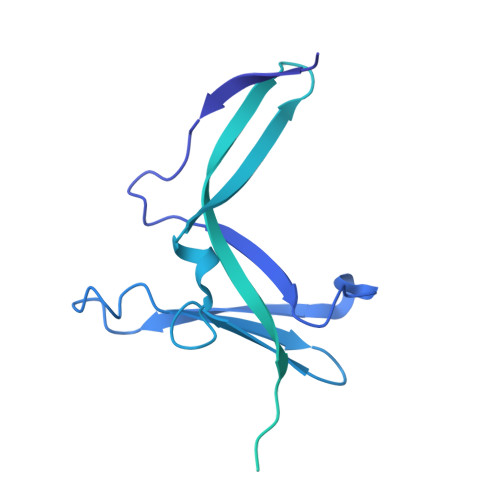
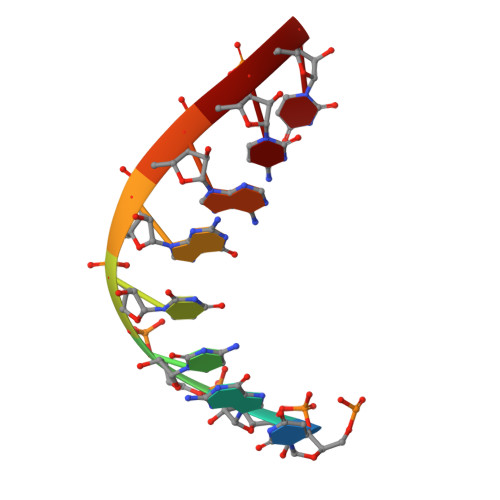
Structure of the human RNA polymerase I elongation complex.
Zhao, D., Liu, W., Chen, K., Wu, Z., Yang, H., Xu, Y.(2021) Cell Discov 7: 97-97
- PubMed: 34671025
- DOI: https://doi.org/10.1038/s41421-021-00335-5
- Primary Citation of Related Structures:
7VBA, 7VBB, 7VBC - PubMed Abstract:
Eukaryotic RNA polymerase I (Pol I) transcribes ribosomal DNA and generates RNA for ribosome synthesis. Pol I accounts for the majority of cellular transcription activity and dysregulation of Pol I transcription leads to cancers and ribosomopathies. Despite extensive structural studies of yeast Pol I, structure of human Pol I remains unsolved. Here we determined the structures of the human Pol I in the pre-translocation, post-translocation, and backtracked states at near-atomic resolution. The single-subunit peripheral stalk lacks contacts with the DNA-binding clamp and is more flexible than the two-subunit stalk in yeast Pol I. Compared to yeast Pol I, human Pol I possesses a more closed clamp, which makes more contacts with DNA. The Pol I structure in the post-cleavage backtracked state shows that the C-terminal zinc ribbon of RPA12 inserts into an open funnel and facilitates "dinucleotide cleavage" on mismatched DNA-RNA hybrid. Critical disease-associated mutations are mapped on Pol I regions that are involved in catalysis and complex organization. In summary, the structures provide new sights into human Pol I complex organization and efficient proofreading.
- Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, State Key Laboratory of Genetic Engineering, Shanghai Key Laboratory of Radiation Oncology, and Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College of Fudan University, Shanghai, China.
Organizational Affiliation: