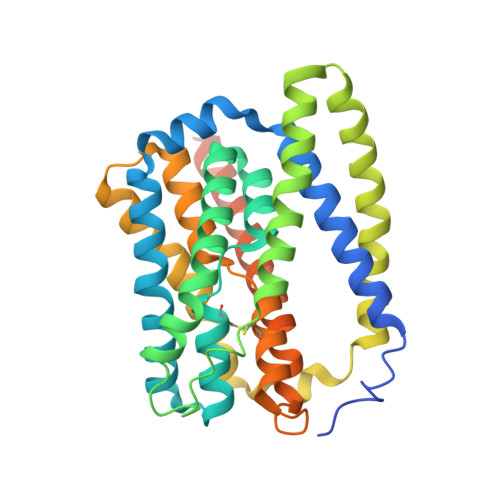
Structure of the bile acid transporter and HBV receptor NTCP.
Asami, J., Kimura, K.T., Fujita-Fujiharu, Y., Ishida, H., Zhang, Z., Nomura, Y., Liu, K., Uemura, T., Sato, Y., Ono, M., Yamamoto, M., Noda, T., Shigematsu, H., Drew, D., Iwata, S., Shimizu, T., Nomura, N., Ohto, U.(2022) Nature 606: 1021-1026
- PubMed: 35580629
- DOI: https://doi.org/10.1038/s41586-022-04845-4
- Primary Citation of Related Structures:
7VAD, 7VAE, 7VAF, 7VAG, 7WSI - PubMed Abstract:
Chronic infection with hepatitis B virus (HBV) affects more than 290 million people worldwide, is a major cause of cirrhosis and hepatocellular carcinoma, and results in an estimated 820,000 deaths annually 1,2 . For HBV infection to be established, a molecular interaction is required between the large glycoproteins of the virus envelope (known as LHBs) and the host entry receptor sodium taurocholate co-transporting polypeptide (NTCP), a sodium-dependent bile acid transporter from the blood to hepatocytes 3 . However, the molecular basis for the virus-transporter interaction is poorly understood. Here we report the cryo-electron microscopy structures of human, bovine and rat NTCPs in the apo state, which reveal the presence of a tunnel across the membrane and a possible transport route for the substrate. Moreover, the cryo-electron microscopy structure of human NTCP in the presence of the myristoylated preS1 domain of LHBs, together with mutation and transport assays, suggest a binding mode in which preS1 and the substrate compete for the extracellular opening of the tunnel in NTCP. Our preS1 domain interaction analysis enables a mechanistic interpretation of naturally occurring HBV-insusceptible mutations in human NTCP. Together, our findings provide a structural framework for HBV recognition and a mechanistic understanding of sodium-dependent bile acid translocation by mammalian NTCPs.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: