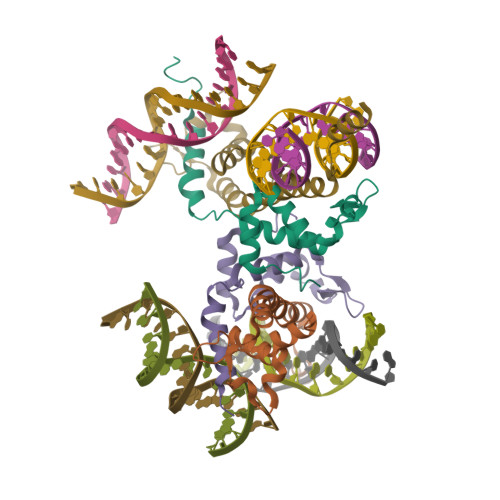
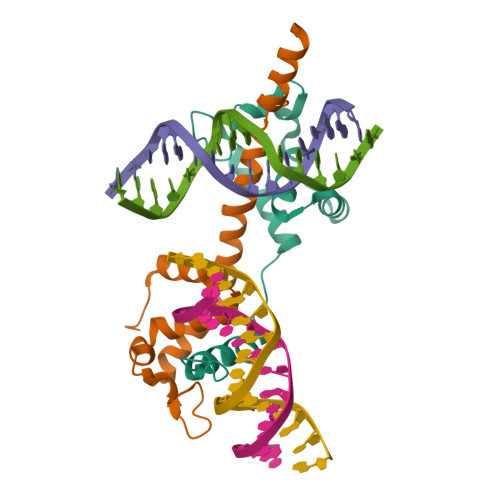
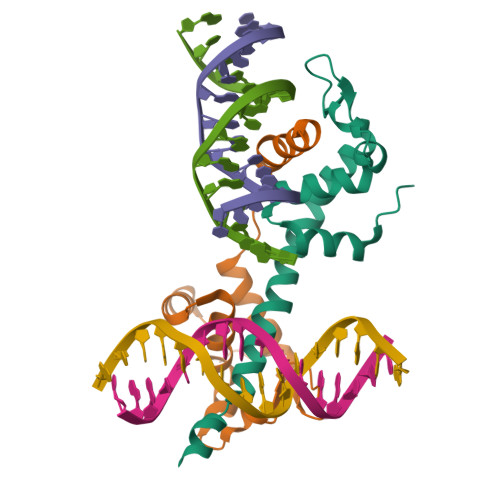
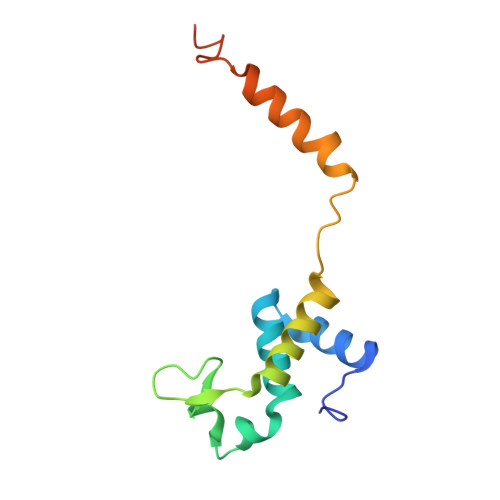
Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Zhang, J., Zhang, Y., You, Q., Huang, C., Zhang, T., Wang, M., Zhang, T., Yang, X., Xiong, J., Li, Y., Liu, C.P., Zhang, Z., Xu, R.M., Zhu, B.(2022) Science 375: 1053-1058
- PubMed: 35143257
- DOI: https://doi.org/10.1126/science.abm0730
- Primary Citation of Related Structures:
7V9F, 7V9G, 7V9H, 7V9I - PubMed Abstract:
Bivalent genes are ready for activation upon the arrival of developmental cues. Here, we report that BEND3 is a CpG island (CGI)-binding protein that is enriched at regulatory elements. The cocrystal structure of BEND3 in complex with its target DNA reveals the structural basis for its DNA methylation-sensitive binding property. Mouse embryos ablated of Bend3 died at the pregastrulation stage. Bend3 null embryonic stem cells (ESCs) exhibited severe defects in differentiation, during which hundreds of CGI-containing bivalent genes were prematurely activated. BEND3 is required for the stable association of polycomb repressive complex 2 (PRC2) at bivalent genes that are highly occupied by BEND3, which suggests a reining function of BEND3 in maintaining high levels of H3K27me3 at these bivalent genes in ESCs to prevent their premature activation in the forthcoming developmental stage.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.