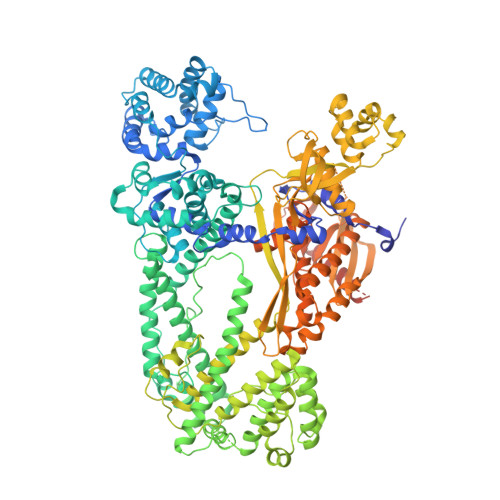
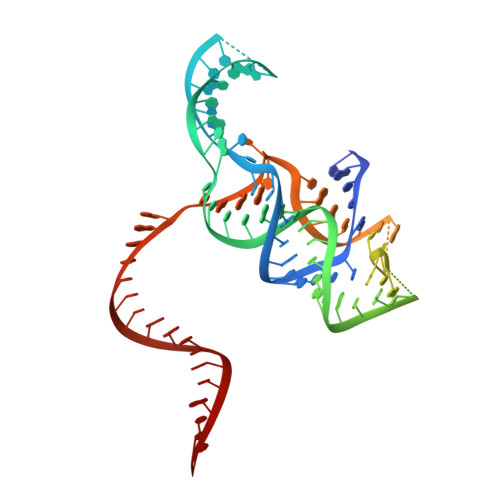
Structure of the type V-C CRISPR-Cas effector enzyme.
Kurihara, N., Nakagawa, R., Hirano, H., Okazaki, S., Tomita, A., Kobayashi, K., Kusakizako, T., Nishizawa, T., Yamashita, K., Scott, D.A., Nishimasu, H., Nureki, O.(2022) Mol Cell 82: 1865-1877.e4
- PubMed: 35366394
- DOI: https://doi.org/10.1016/j.molcel.2022.03.006
- Primary Citation of Related Structures:
7V93, 7V94 - PubMed Abstract:
RNA-guided CRISPR-Cas nucleases are widely used as versatile genome-engineering tools. Recent studies identified functionally divergent type V Cas12 family enzymes. Among them, Cas12c2 binds a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) and recognizes double-stranded DNA targets with a short TN PAM. Here, we report the cryo-electron microscopy structures of the Cas12c2-guide RNA binary complex and the Cas12c2-guide RNA-target DNA ternary complex. The structures revealed that the crRNA and tracrRNA form an unexpected X-junction architecture, and that Cas12c2 recognizes a single T nucleotide in the PAM through specific hydrogen-bonding interactions with two arginine residues. Furthermore, our biochemical analyses indicated that Cas12c2 processes its precursor crRNA to a mature crRNA using the RuvC catalytic site through a unique mechanism. Collectively, our findings improve the mechanistic understanding of diverse type V CRISPR-Cas effectors.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
Organizational Affiliation: