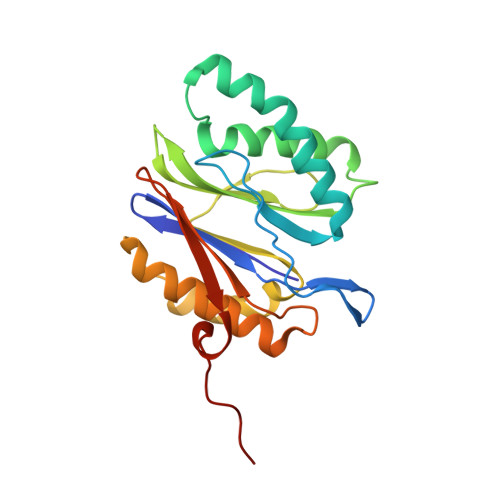
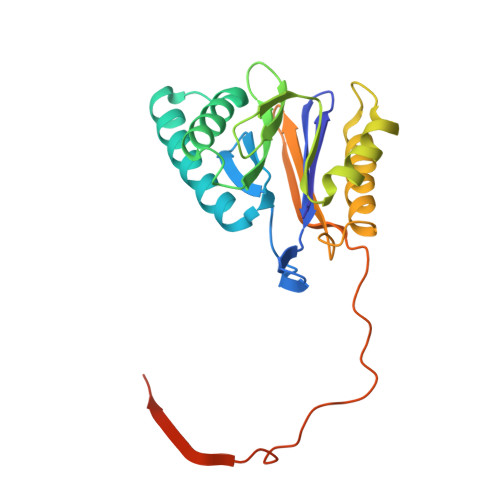
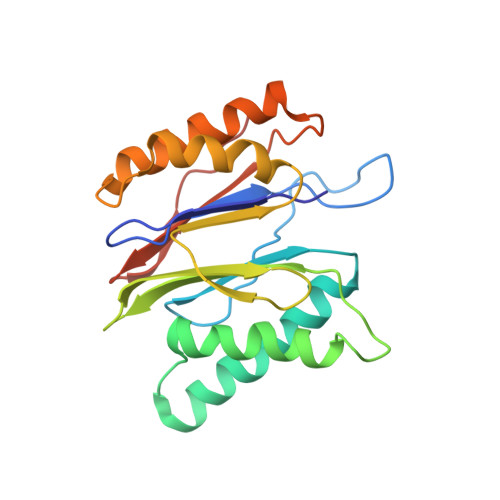
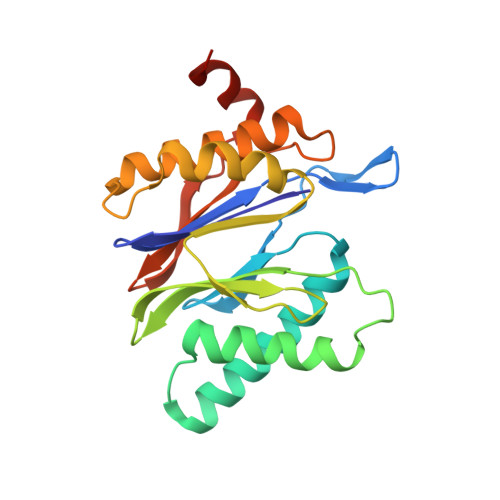
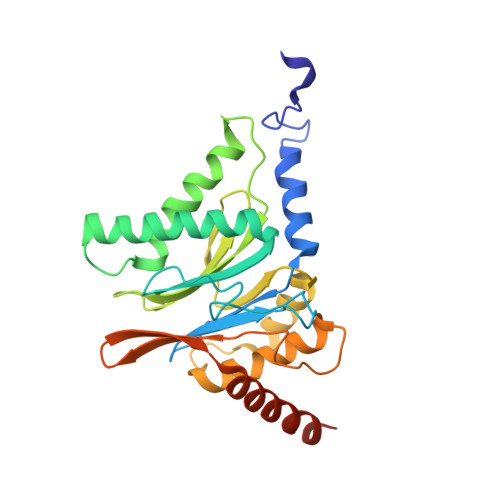
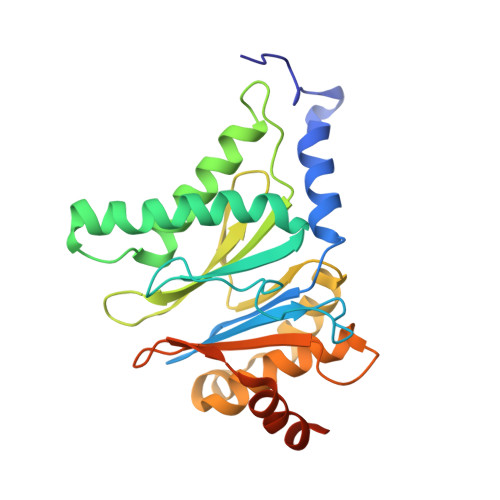
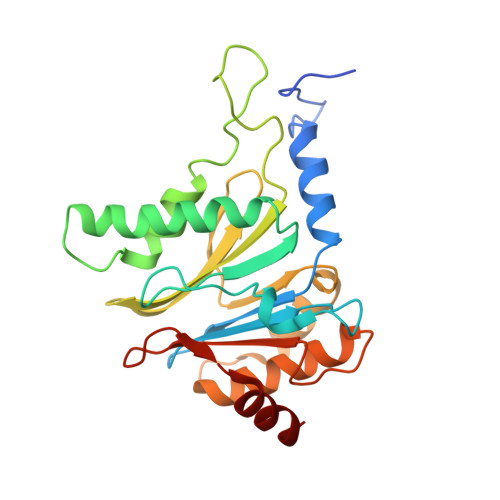
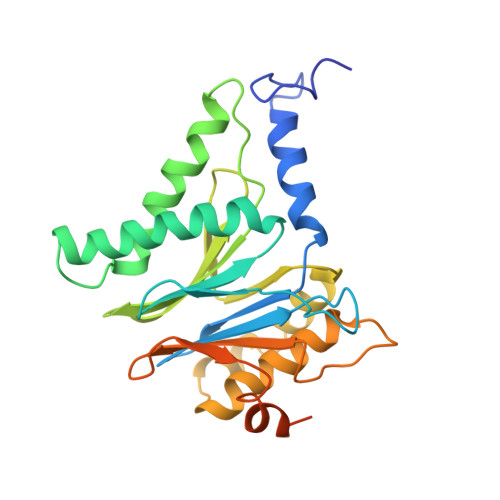
The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Sahu, I., Mali, S.M., Sulkshane, P., Xu, C., Rozenberg, A., Morag, R., Sahoo, M.P., Singh, S.K., Ding, Z., Wang, Y., Day, S., Cong, Y., Kleifeld, O., Brik, A., Glickman, M.H.(2021) Nat Commun 12: 6173-6173
- PubMed: 34702852
- DOI: https://doi.org/10.1038/s41467-021-26427-0
- Primary Citation of Related Structures:
7PG9, 7V5G, 7V5M - PubMed Abstract:
The proteasome, the primary protease for ubiquitin-dependent proteolysis in eukaryotes, is usually found as a mixture of 30S, 26S, and 20S complexes. These complexes have common catalytic sites, which makes it challenging to determine their distinctive roles in intracellular proteolysis. Here, we chemically synthesize a panel of homogenous ubiquitinated proteins, and use them to compare 20S and 26S proteasomes with respect to substrate selection and peptide-product generation. We show that 20S proteasomes can degrade the ubiquitin tag along with the conjugated substrate. Ubiquitin remnants on branched peptide products identified by LC-MS/MS, and flexibility in the 20S gate observed by cryo-EM, reflect the ability of the 20S proteasome to proteolyze an isopeptide-linked ubiquitin-conjugate. Peptidomics identifies proteasome-trapped ubiquitin-derived peptides and peptides of potential 20S substrates in Hi20S cells, hypoxic cells, and human failing-heart. Moreover, elevated levels of 20S proteasomes appear to contribute to cell survival under stress associated with damaged proteins.
- Faculty of Biology, Technion-Israel Institute of Technology, Haifa, 32000, Israel.
Organizational Affiliation: