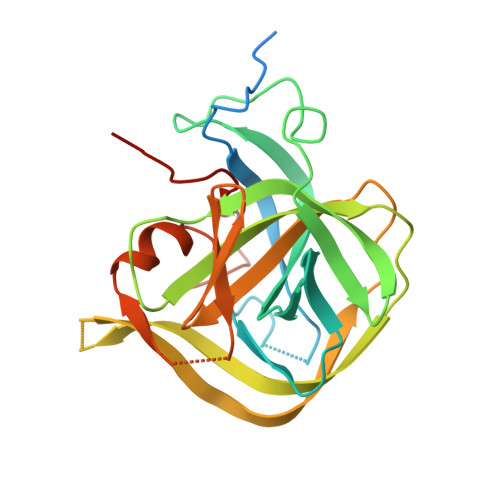
Structure of the divergent human astrovirus MLB capsid spike.
Delgado-Cunningham, K., Lopez, T., Khatib, F., Arias, C.F., DuBois, R.M.(2022) Structure 30: 1573-1581.e3
- PubMed: 36417907
- DOI: https://doi.org/10.1016/j.str.2022.10.010
- Primary Citation of Related Structures:
7UZT - PubMed Abstract:
Despite their worldwide prevalence and association with human disease, the molecular bases of human astrovirus (HAstV) infection and evolution remain poorly characterized. Here, we report the structure of the capsid protein spike of the divergent HAstV MLB clade (HAstV MLB). While the structure shares a similar folding topology with that of classical-clade HAstV spikes, it is otherwise strikingly different. We find no evidence of a conserved receptor-binding site between the MLB and classical HAstV spikes, suggesting that MLB and classical HAstVs utilize different receptors for host-cell attachment. We provide evidence for this hypothesis using a novel HAstV infection competition assay. Comparisons of the HAstV MLB spike structure with structures predicted from its sequence reveal poor matches, but template-based predictions were surprisingly accurate relative to machine-learning-based predictions. Our data provide a foundation for understanding the mechanisms of infection by diverse HAstVs and can support structure determination in similarly unstudied systems.
- Department of Biomolecular Engineering, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.
Organizational Affiliation: