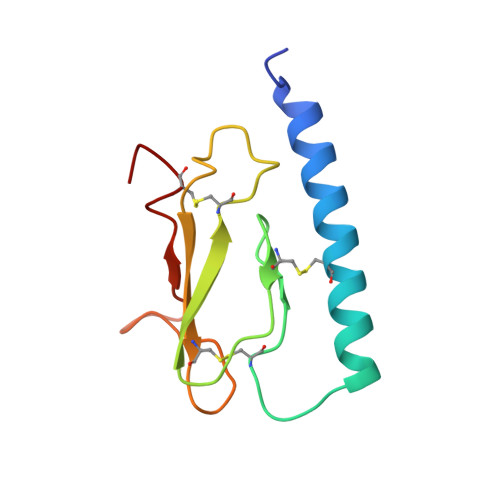
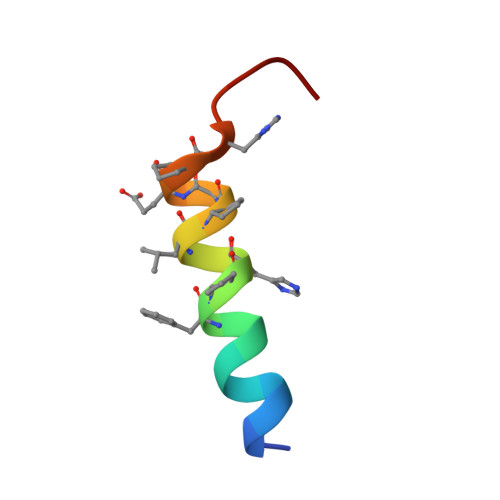
Altered signaling at the PTH receptor via modified agonist contacts with the extracellular domain provides a path to prolonged agonism in vivo.
Liu, S., Yu, Z., Daley, E.J., Bingman, C.A., Bruchs, A.T., Gardella, T.J., Gellman, S.H.(2022) Proc Natl Acad Sci U S A 119: e2212736119-e2212736119
- PubMed: 36409914
- DOI: https://doi.org/10.1073/pnas.2212736119
- Primary Citation of Related Structures:
7UZO, 7UZP - PubMed Abstract:
The parathyroid hormone type 1 receptor (PTHR1), a Class B GPCR, is activated by long polypeptides, including drugs for osteoporosis and hypoparathyroidism. The PTHR1 engages peptide agonists via a two-step mechanism. Initial contact involves the extracellular domain (ECD), which has been thought to contribute primarily to receptor-peptide binding, and then the N terminus of the peptide engages the receptor transmembrane domain (TMD), which is thought to control the message conveyed to intracellular partners. This mechanism has been suggested to apply to other Class B GPCRs as well. Here, we show that modification of a PTHR1 agonist at ECD-contact sites can alter the signaling profile, an outcome that is not accommodated by the current two-step binding model. Our data support a modified two-step binding model in which agonist orientation on the ECD surface can influence the geometry of agonist-TMD engagement. This expanded binding model offers a mechanism by which altering ECD-contact residues can affect signaling profile. Our discoveries provide a rationale for exploring agonist modifications distal from the TMD-contact region in future efforts to optimize therapeutic performance of peptide hormone analogs.
- Department of Chemistry, University of Wisconsin, Madison, WI 53706.
Organizational Affiliation: