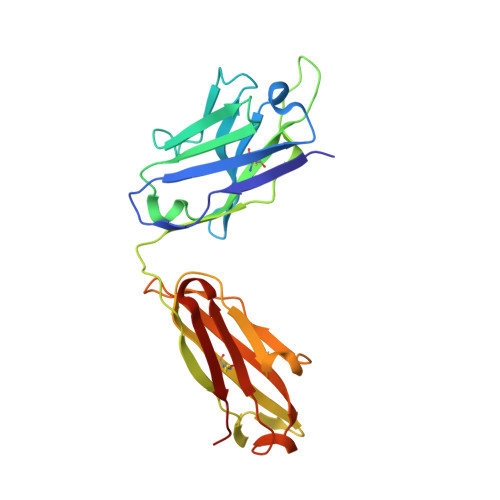
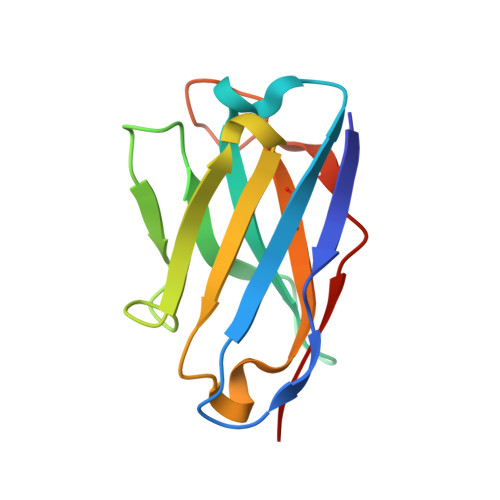
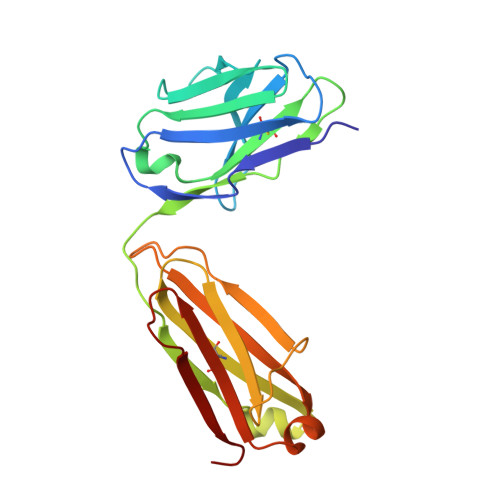
High-density binding to Plasmodium falciparum circumsporozoite protein repeats by inhibitory antibody elicited in mouse with human immunoglobulin repertoire.
Kucharska, I., Binter, S., Murugan, R., Scally, S.W., Ludwig, J., Prieto, K., Thai, E., Costa, G., Li, K., Horn, G.Q., Flores-Garcia, Y., Bosch, A., Sicard, T., Rubinstein, J.L., Zavala, F., Dennison, S.M., Tomaras, G.D., Levashina, E.A., Kellam, P., Wardemann, H., Julien, J.P.(2022) PLoS Pathog 18: e1010999-e1010999
- PubMed: 36441829
- DOI: https://doi.org/10.1371/journal.ppat.1010999
- Primary Citation of Related Structures:
7UYL, 7UYM, 7V05 - PubMed Abstract:
Antibodies targeting the human malaria parasite Plasmodium falciparum circumsporozoite protein (PfCSP) can prevent infection and disease. PfCSP contains multiple central repeating NANP motifs; some of the most potent anti-infective antibodies against malaria bind to these repeats. Multiple antibodies can bind the repeating epitopes concurrently by engaging into homotypic Fab-Fab interactions, which results in the ordering of the otherwise largely disordered central repeat into a spiral. Here, we characterize IGHV3-33/IGKV1-5-encoded monoclonal antibody (mAb) 850 elicited by immunization of transgenic mice with human immunoglobulin loci. mAb 850 binds repeating NANP motifs with picomolar affinity, potently inhibits Plasmodium falciparum (Pf) in vitro and, when passively administered in a mouse challenge model, reduces liver burden to a similar extent as some of the most potent anti-PfCSP mAbs yet described. Like other IGHV3-33/IGKV1-5-encoded anti-NANP antibodies, mAb 850 primarily utilizes its HCDR3 and germline-encoded aromatic residues to recognize its core NANP motif. Biophysical and cryo-electron microscopy analyses reveal that up to 19 copies of Fab 850 can bind the PfCSP repeat simultaneously, and extensive homotypic interactions are observed between densely-packed PfCSP-bound Fabs to indirectly improve affinity to the antigen. Together, our study expands on the molecular understanding of repeat-induced homotypic interactions in the B cell response against PfCSP for potently protective mAbs against Pf infection.
- Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, Canada.
Organizational Affiliation: