Uncovering translation roadblocks during the development of a synthetic tRNA.
Prabhakar, A., Krahn, N., Zhang, J., Vargas-Rodriguez, O., Krupkin, M., Fu, Z., Acosta-Reyes, F.J., Ge, X., Choi, J., Crnkovic, A., Ehrenberg, M., Puglisi, E.V., Soll, D., Puglisi, J.(2022) Nucleic Acids Res 50: 10201-10211
- PubMed: 35882385
- DOI: https://doi.org/10.1093/nar/gkac576
- Primary Citation of Related Structures:
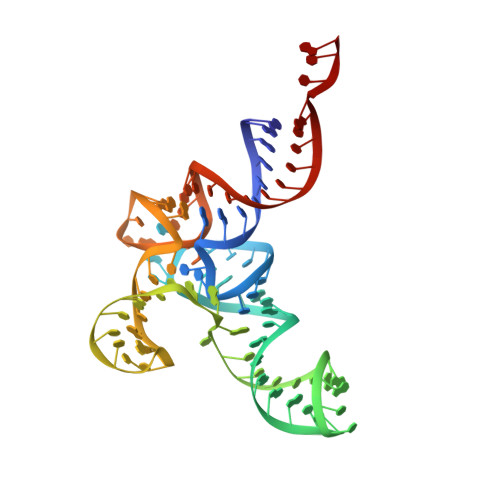
7UR5, 7URI, 7URM - PubMed Abstract:
Ribosomes are remarkable in their malleability to accept diverse aminoacyl-tRNA substrates from both the same organism and other organisms or domains of life. This is a critical feature of the ribosome that allows the use of orthogonal translation systems for genetic code expansion. Optimization of these orthogonal translation systems generally involves focusing on the compatibility of the tRNA, aminoacyl-tRNA synthetase, and a non-canonical amino acid with each other. As we expand the diversity of tRNAs used to include non-canonical structures, the question arises as to the tRNA suitability on the ribosome. Specifically, we investigated the ribosomal translation of allo-tRNAUTu1, a uniquely shaped (9/3) tRNA exploited for site-specific selenocysteine insertion, using single-molecule fluorescence. With this technique we identified ribosomal disassembly occurring from translocation of allo-tRNAUTu1 from the A to the P site. Using cryo-EM to capture the tRNA on the ribosome, we pinpointed a distinct tertiary interaction preventing fluid translocation. Through a single nucleotide mutation, we disrupted this tertiary interaction and relieved the translation roadblock. With the continued diversification of genetic code expansion, our work highlights a targeted approach to optimize translation by distinct tRNAs as they move through the ribosome.
- Department of Structural Biology, Stanford University, Stanford, CA 94305-5126, USA.
Organizational Affiliation: