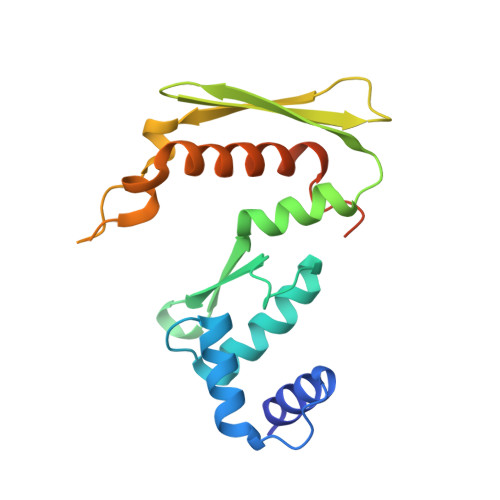
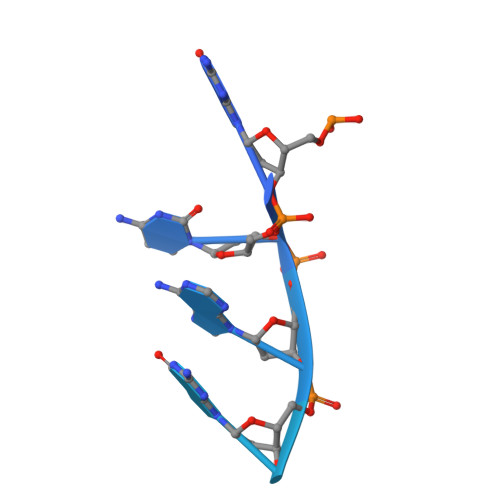
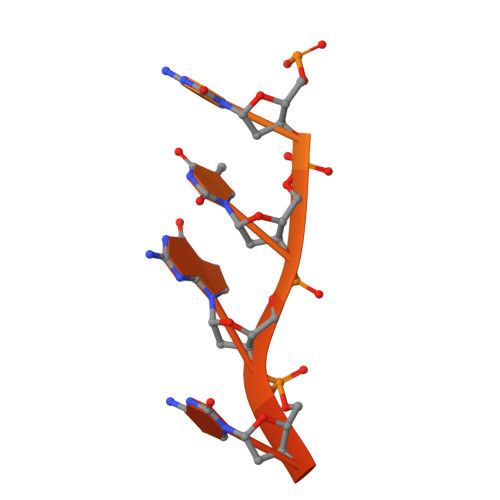
Red beta 177 annealase structure reveals details of oligomerization and lambda Red-mediated homologous DNA recombination
Newing, T.P., Brewster, J.L., Fitschen, L.J., Bouwer, J.C., Johnston, N.P., Yu, H., Tolun, G.(2022) Nat Commun 13: 5649
- PubMed: 36163171
- DOI: https://doi.org/10.1038/s41467-022-33090-6
- Primary Citation of Related Structures:
7UJL - PubMed Abstract:
The Redβ protein of the bacteriophage λ red recombination system is a model annealase which catalyzes single-strand annealing homologous DNA recombination. Here we present the structure of a helical oligomeric annealing intermediate of Redβ, consisting of N-terminal residues 1-177 bound to two complementary 27mer oligonucleotides, determined via cryogenic electron microscopy (cryo-EM) to a final resolution of 3.3 Å. The structure reveals a continuous binding groove which positions and stabilizes complementary DNA strands in a planar orientation to facilitate base pairing via a network of hydrogen bonding. Definition of the inter-subunit interface provides a structural basis for the propensity of Redβ to oligomerize into functionally significant long helical filaments, a trait shared by most annealases. Our cryo-EM structure and molecular dynamics simulations suggest that residues 133-138 form a flexible loop which modulates access to the binding groove. More than half a century after its discovery, this combination of structural and computational observations has allowed us to propose molecular mechanisms for the actions of the model annealase Redβ, a defining member of the Redβ/RecT protein family.
- School of Chemistry and Molecular Bioscience, and Molecular Horizons, University of Wollongong, Wollongong, NSW, Australia.
Organizational Affiliation: