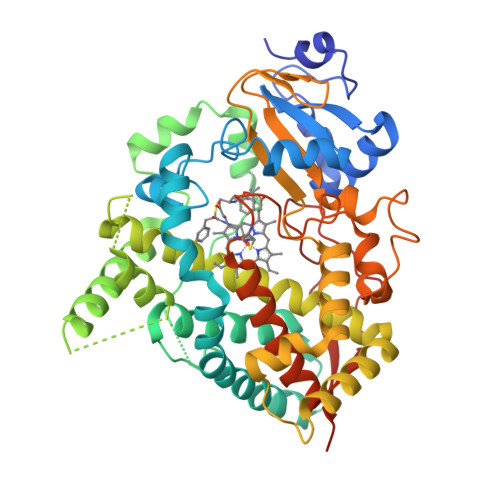
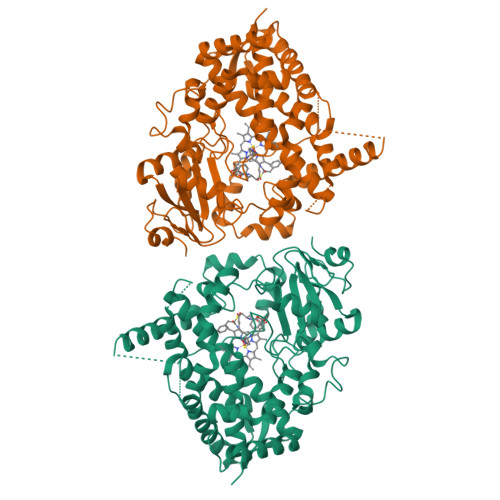
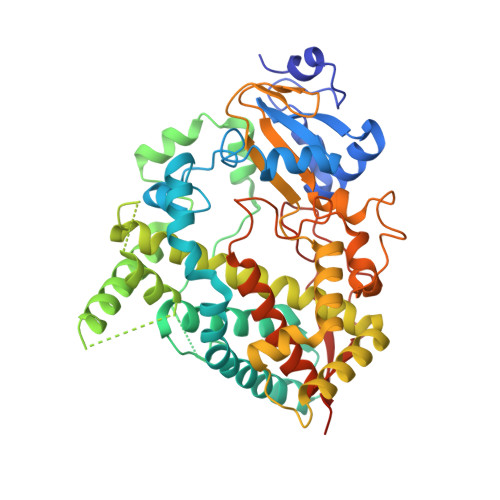
Interaction of CYP3A4 with Rationally Designed Ritonavir Analogues: Impact of Steric Constraints Imposed on the Heme-Ligating Group and the End-Pyridine Attachment.
Samuels, E.R., Sevrioukova, I.F.(2022) Int J Mol Sci 23
- PubMed: 35806297
- DOI: https://doi.org/10.3390/ijms23137291
- Primary Citation of Related Structures:
7UF9, 7UFA, 7UFB, 7UFC, 7UFD, 7UFE, 7UFF - PubMed Abstract:
Controlled inhibition of drug-metabolizing cytochrome P450 3A4 (CYP3A4) is utilized to boost bioavailability of anti-viral and immunosuppressant pharmaceuticals. We investigate structure-activity relationships (SARs) in analogues of ritonavir, a potent CYP3A4 inhibitor marketed as pharmacoenhancer, to determine structural elements required for potent inhibition and whether the inhibitory potency can be further improved via a rational structure-based design. This study investigated eight (series VI) inhibitors differing in head- and end-moieties and their respective linkers. SAR analysis revealed the multifactorial regulation of inhibitory strength, with steric constraints imposed on the tethered heme-ligating moiety being a key factor. Minimization of these constraints by changing the linkers' length/flexibility and N-heteroatom position strengthened heme coordination and markedly improved binding and/or inhibitory strength. Impact of the end-pyridine attachment was not uniform due to influence of other determinants controlling the ligand-binding mode. This interplay between pharmacophoric determinants and the end-group enlargement can be used for further inhibitor optimization.
Organizational Affiliation:
Department of Pharmaceutical Sciences, University of California, Irvine, CA 92697, USA.