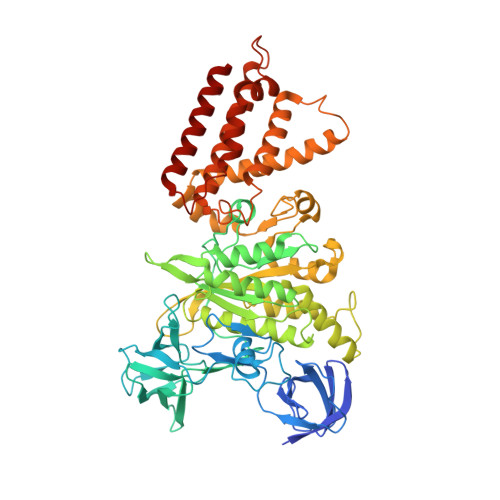
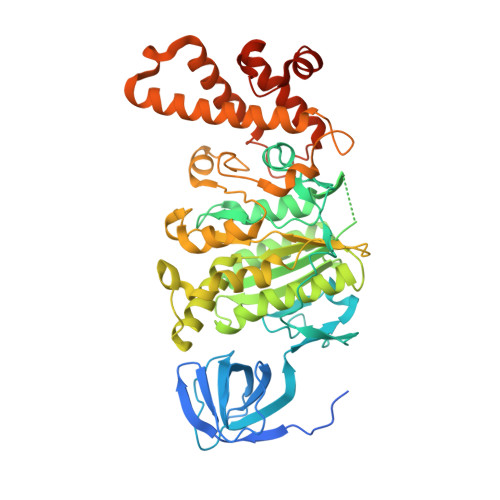
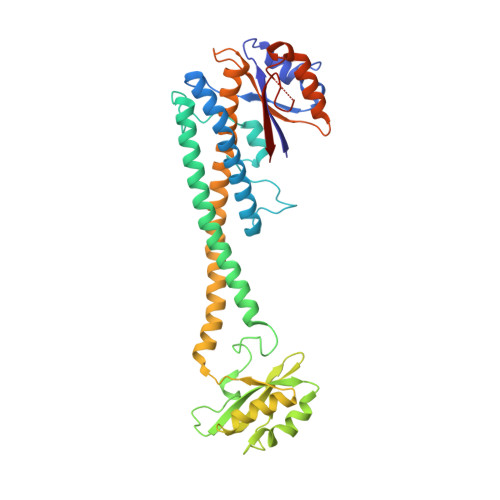
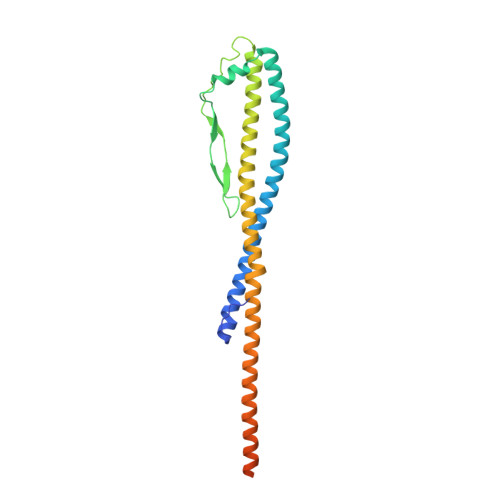
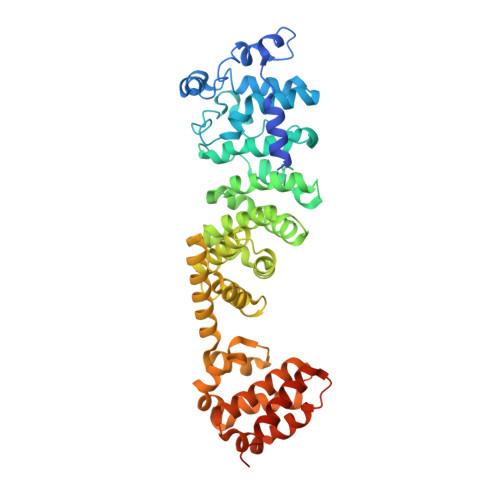
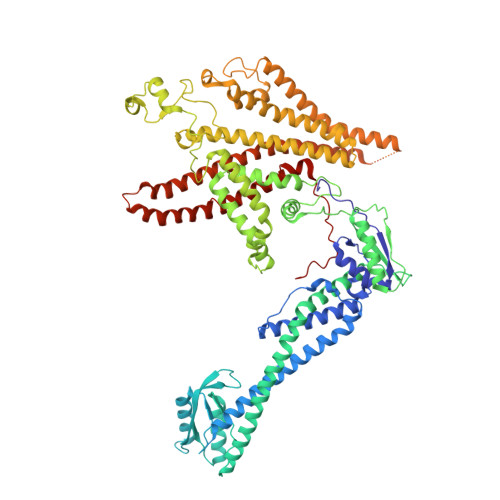
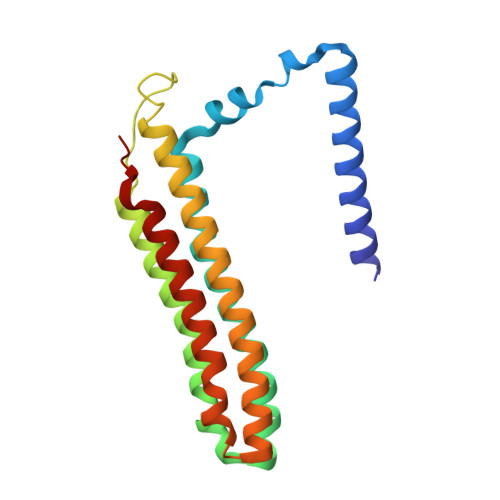
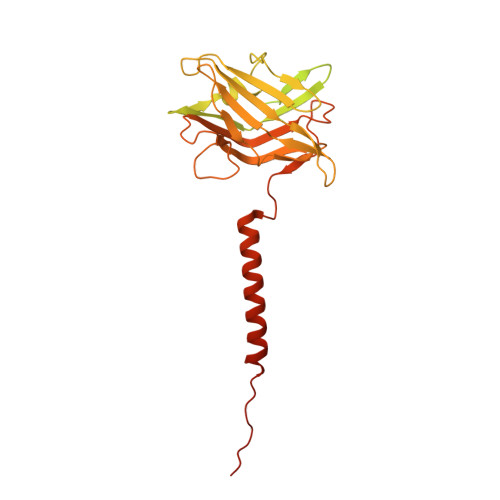
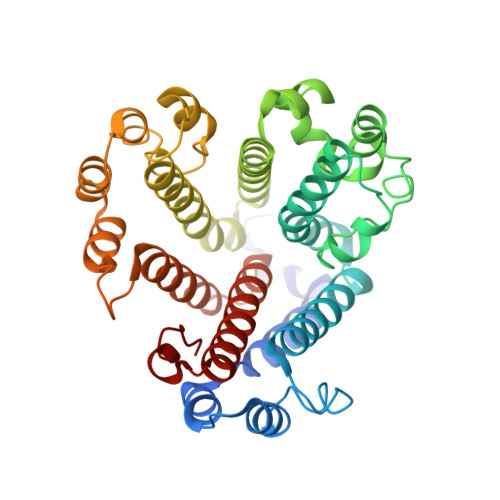
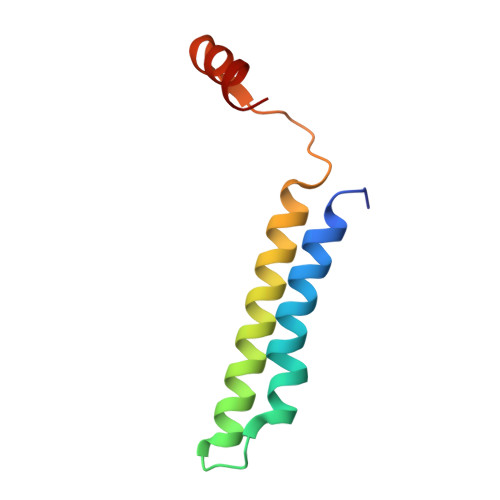
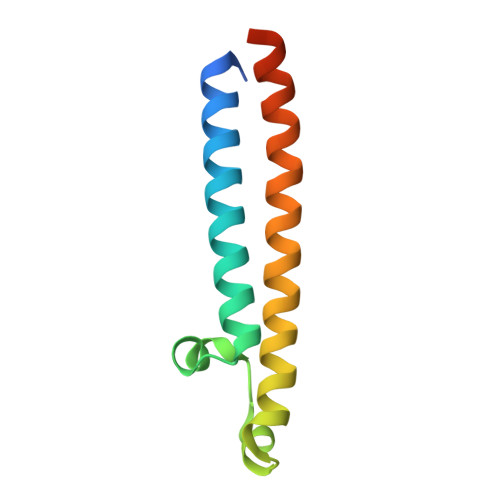
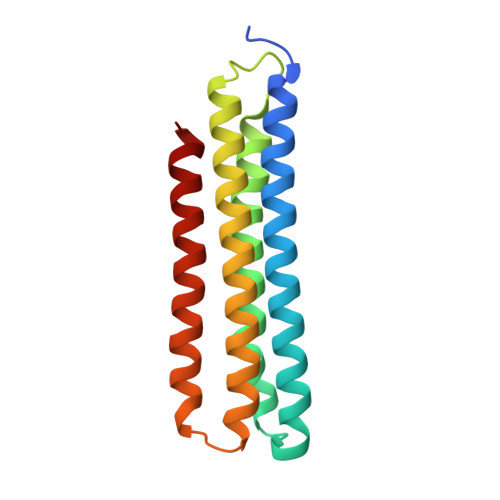
CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Tan, Y.Z., Abbas, Y.M., Wu, J.Z., Wu, D., Keon, K.A., Hesketh, G.G., Bueler, S.A., Gingras, A.C., Robinson, C.V., Grinstein, S., Rubinstein, J.L.(2022) Life Sci Alliance 5
- PubMed: 35794005
- DOI: https://doi.org/10.26508/lsa.202201527
- Primary Citation of Related Structures:
7U8O, 7U8P, 7U8Q, 7U8R - PubMed Abstract:
V-ATPases are rotary proton pumps that serve as signaling hubs with numerous protein binding partners. CryoEM with exhaustive focused classification allowed detection of endogenous proteins associated with porcine kidney V-ATPase. An extra C subunit was found in ∼3% of complexes, whereas ∼1.6% of complexes bound mEAK-7, a protein with proposed roles in dauer formation in nematodes and mTOR signaling in mammals. High-resolution cryoEM of porcine kidney V-ATPase with recombinant mEAK-7 showed that mEAK-7's TLDc domain interacts with V-ATPase's stator, whereas its C-terminal α helix binds V-ATPase's rotor. This crosslink would be expected to inhibit rotary catalysis. However, unlike the yeast TLDc protein Oxr1p, exogenous mEAK-7 does not inhibit V-ATPase and mEAK-7 overexpression in cells does not alter lysosomal or phagosomal pH. Instead, cryoEM suggests that the mEAK-7:V-ATPase interaction is disrupted by ATP-induced rotation of the rotor. Comparison of Oxr1p and mEAK-7 binding explains this difference. These results show that V-ATPase binding by TLDc domain proteins can lead to effects ranging from strong inhibition to formation of labile interactions that are sensitive to the enzyme's activity.
- Molecular Medicine Program, The Hospital for Sick Children Research Institute, Toronto, Canada.
Organizational Affiliation: