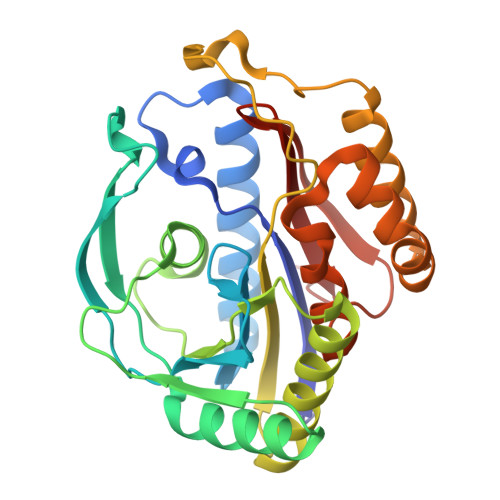
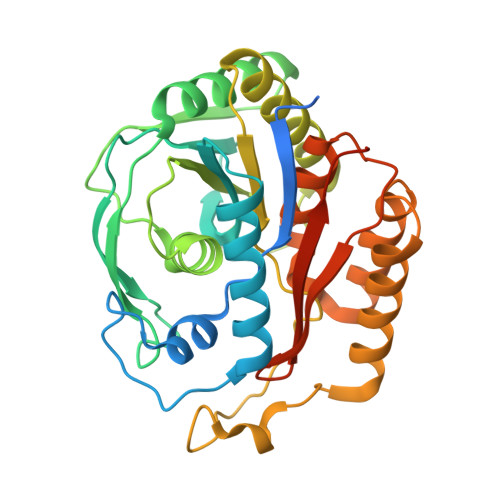
Characterization of a novel aromatic substrate-processing microcompartment in Actinobacteria.
Doron, L., Sutter, M., Kerfeld, C.A.(2023) mBio 14: e0121623-e0121623
- PubMed: 37462359
- DOI: https://doi.org/10.1128/mbio.01216-23
- Primary Citation of Related Structures:
7TXY - PubMed Abstract:
We have discovered a new cluster of genes that is found exclusively in the Actinobacteria phylum. This locus includes genes for the 2-aminophenol meta -cleavage pathway and the shell proteins of a bacterial microcompartment (BMC) and has been named aromatics (ARO) for its putative role in the breakdown of aromatic compounds. In this study, we provide details about the distribution and composition of the ARO BMC locus and conduct phylogenetic, structural, and functional analyses of the first two enzymes in the catabolic pathway: a unique 2-aminophenol dioxygenase, which is exclusively found alongside BMC shell genes in Actinobacteria, and a semialdehyde dehydrogenase, which works downstream of the dioxygenase. Genomic analysis reveals variations in the complexity of the ARO loci across different orders. Some loci are simple, containing shell proteins and enzymes for the initial steps of the catabolic pathway, while others are extensive, encompassing all the necessary genes for the complete breakdown of 2-aminophenol into pyruvate and acetyl-CoA. Furthermore, our analysis uncovers two subtypes of ARO BMC that likely degrade either 2-aminophenol or catechol, depending on the presence of a pathway-specific gene within the ARO locus. The precise precursor of 2-aminophenol, which serves as the initial substrate and/or inducer for the ARO pathway, remains unknown, as our model organism Micromonospora rosaria cannot utilize 2-aminophenol as its sole energy source. However, using enzymatic assays, we demonstrate the dioxygenase's ability to cleave both 2-aminophenol and catechol in vitro , in collaboration with the aldehyde dehydrogenase, to facilitate the rapid conversion of these unstable and toxic intermediates. IMPORTANCE Bacterial microcompartments (BMCs) are proteinaceous organelles that are widespread among bacteria and provide a competitive advantage in specific environmental niches. Studies have shown that the genetic information necessary to form functional BMCs is encoded in loci that contain genes encoding shell proteins and the enzymatic core. This allows the bioinformatic discovery of BMCs with novel functions and expands our understanding of the metabolic diversity of BMCs. ARO loci, found only in Actinobacteria, contain genes encoding for phylogenetically remote shell proteins and homologs of the meta -cleavage degradation pathway enzymes that were shown to convert central aromatic intermediates into pyruvate and acetyl-CoA in gamma Proteobacteria. By analyzing the gene composition of ARO BMC loci and characterizing two core enzymes phylogenetically, structurally, and functionally, we provide an initial functional characterization of the ARO BMC, the most unusual BMC identified to date, distinctive among the repertoire of studied BMCs.
Organizational Affiliation:
MSU-DOE Plant Research Laboratory, Michigan State University , East Lansing, Michigan, USA.