Structure of C. albicans FAS in an inhibited state.
Mazhab-Jafari, M.T., Lou, J.W.Not Published
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fatty acid synthase subunit alpha | 1,885 | Candida albicans | Mutation(s): 0 EC: 2.3.1.86 (PDB Primary Data), 1.1.1.100 (PDB Primary Data), 2.3.1.41 (PDB Primary Data) | 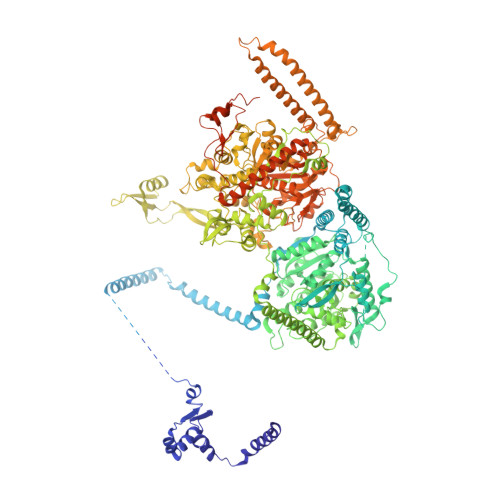 | |
UniProt | |||||
Find proteins for P43098 (Candida albicans) Explore P43098 Go to UniProtKB: P43098 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P43098 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fatty acid synthase subunit beta | 2,037 | Candida albicans | Mutation(s): 0 EC: 2.3.1.86 (PDB Primary Data), 4.2.1.59 (PDB Primary Data), 1.3.1.9 (PDB Primary Data), 2.3.1.38 (PDB Primary Data), 2.3.1.39 (PDB Primary Data), 3.1.2.14 (PDB Primary Data) | 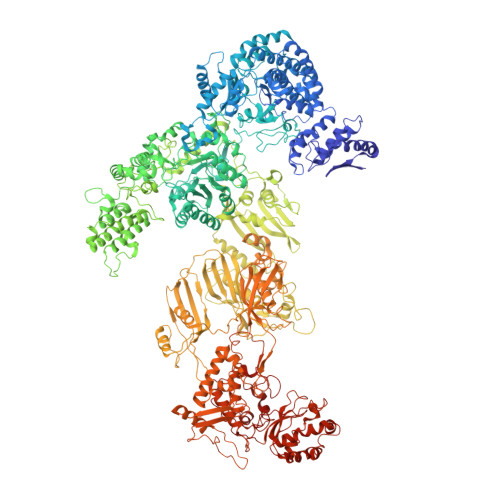 | |
UniProt | |||||
Find proteins for P34731 (Candida albicans) Explore P34731 Go to UniProtKB: P34731 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P34731 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FMN (Subject of Investigation/LOI) Query on FMN | C [auth B] | FLAVIN MONONUCLEOTIDE C17 H21 N4 O9 P FVTCRASFADXXNN-SCRDCRAPSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 3 |
| MODEL REFINEMENT | PHENIX | |
| MODEL REFINEMENT | Coot |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | -- |