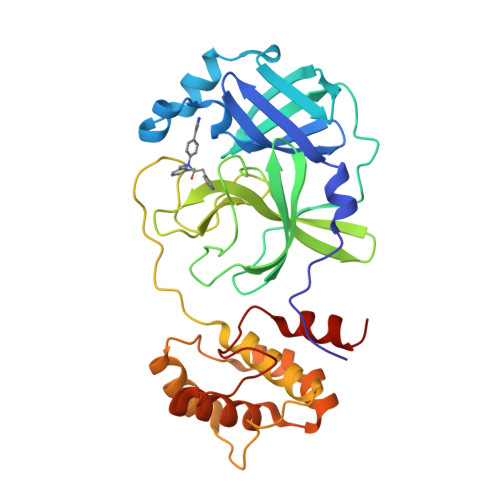
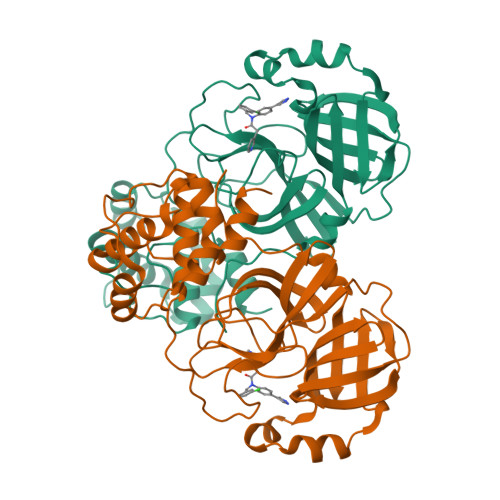
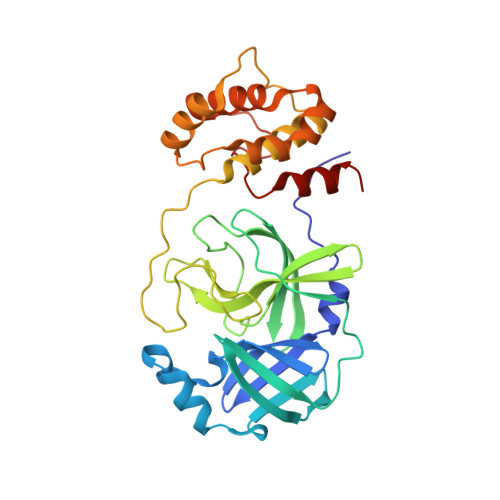
SARS-CoV-2 3CL-protease inhibitors derived from ML300: investigation of P1 and replacements of the 1,2,3-benzotriazole
Hooper, A., Macdonald, J.D., Reilly, B., Maw, J., Wirrick, A.P., Han, S.H., Lindsey, A.A., Rico, E.G., Romigh, T., Goins, C.M., Wang, N.S., Stauffer, S.R.(2023) Med Chem Res