Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Zhang, K., Horikoshi, N., Li, S., Powers, A.S., Hameedi, M.A., Pintilie, G.D., Chae, H.D., Khan, Y.A., Suomivuori, C.M., Dror, R.O., Sakamoto, K.M., Chiu, W., Wakatsuki, S.(2022) ACS Cent Sci 8: 214-222
- PubMed: 35233453
- DOI: https://doi.org/10.1021/acscentsci.1c01090
- Primary Citation of Related Structures:
7TB3, 7TBH - PubMed Abstract:
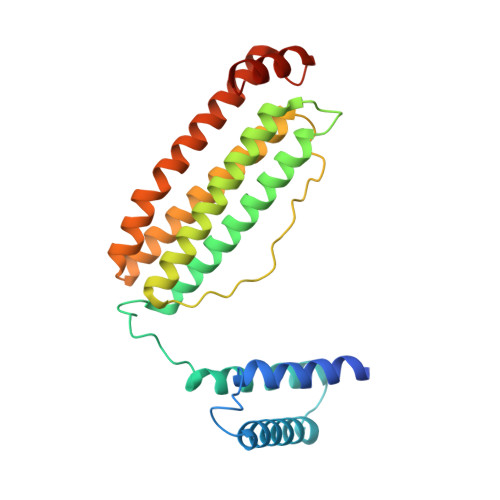
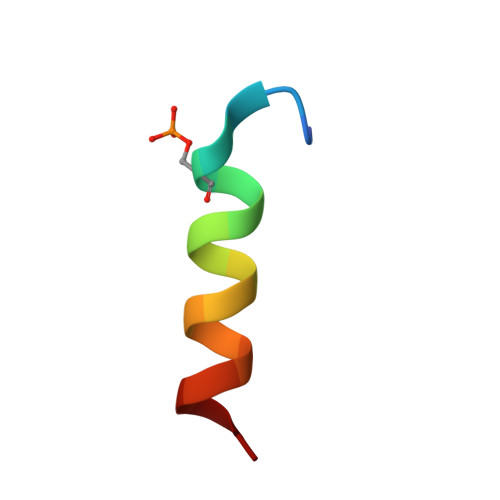
Cryogenic electron microscopy (cryo-EM) has emerged as a viable structural tool for molecular therapeutics development against human diseases. However, it remains a challenge to determine structures of proteins that are flexible and smaller than 30 kDa. The 11 kDa KIX domain of CREB-binding protein (CBP), a potential therapeutic target for acute myeloid leukemia and other cancers, is a protein which has defied structure-based inhibitor design. Here, we develop an experimental approach to overcome the size limitation by engineering a protein double-shell to sandwich the KIX domain between apoferritin as the inner shell and maltose-binding protein as the outer shell. To assist homogeneous orientations of the target, disulfide bonds are introduced at the target-apoferritin interface, resulting in a cryo-EM structure at 2.6 Å resolution. We used molecular dynamics simulations to design peptides that block the interaction of the KIX domain of CBP with the intrinsically disordered pKID domain of CREB. The double-shell design allows for fluorescence polarization assays confirming the binding between the KIX domain in the double-shell and these interacting peptides. Further cryo-EM analysis reveals a helix-helix interaction between a single KIX helix and the best peptide, providing a possible strategy for developments of next-generation inhibitors.
- MOE Key Laboratory for Cellular Dynamics and Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, China.
Organizational Affiliation: