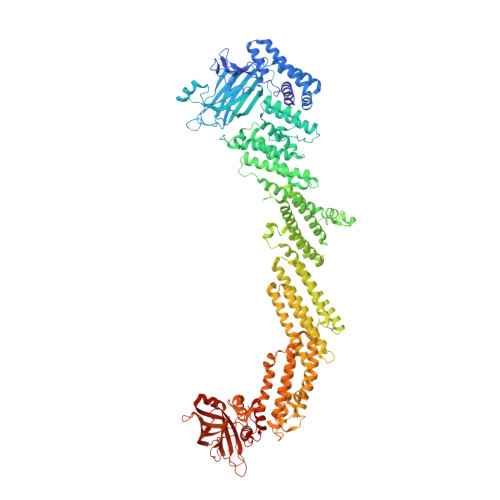
Munc13 structural transitions and oligomers that may choreograph successive stages in vesicle priming for neurotransmitter release.
Grushin, K., Kalyana Sundaram, R.V., Sindelar, C.V., Rothman, J.E.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35135883
- DOI: https://doi.org/10.1073/pnas.2121259119
- Primary Citation of Related Structures:
7T7C, 7T7R, 7T7V, 7T7X, 7T81 - PubMed Abstract:
How can exactly six SNARE complexes be assembled under each synaptic vesicle? Here we report cryo-EM crystal structures of the core domain of Munc13, the key chaperone that initiates SNAREpin assembly. The functional core of Munc13, consisting of C1-C2B-MUN-C2C (Munc13C) spontaneously crystallizes between phosphatidylserine-rich bilayers in two distinct conformations, each in a radically different oligomeric state. In the open conformation (state 1), Munc13C forms upright trimers that link the two bilayers, separating them by ∼21 nm. In the closed conformation, six copies of Munc13C interact to form a lateral hexamer elevated ∼14 nm above the bilayer. Open and closed conformations differ only by a rigid body rotation around a flexible hinge, which when performed cooperatively assembles Munc13 into a lateral hexamer (state 2) in which the key SNARE assembly-activating site of Munc13 is autoinhibited by its neighbor. We propose that each Munc13 in the lateral hexamer ultimately assembles a single SNAREpin, explaining how only and exactly six SNARE complexes are templated. We suggest that state 1 and state 2 may represent two successive states in the synaptic vesicle supply chain leading to "primed" ready-release vesicles in which SNAREpins are clamped and ready to release (state 3).
- Department of Cell Biology, Yale University, New Haven, CT 06520.
Organizational Affiliation: