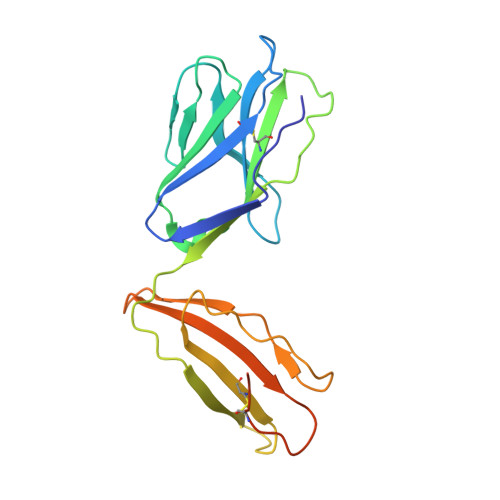
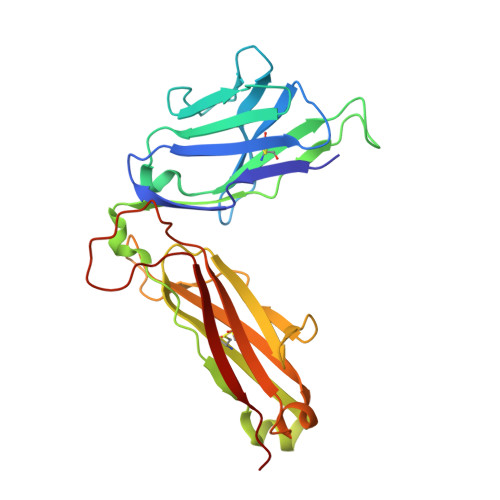
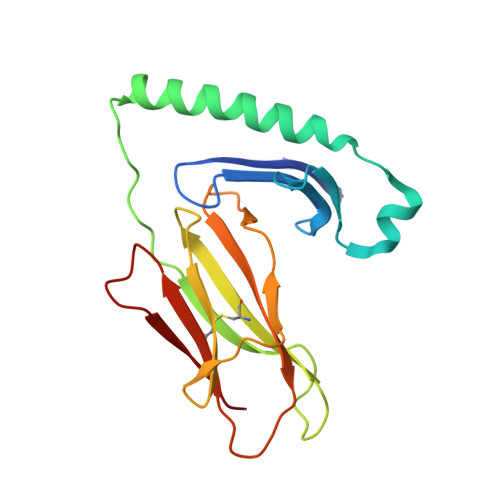
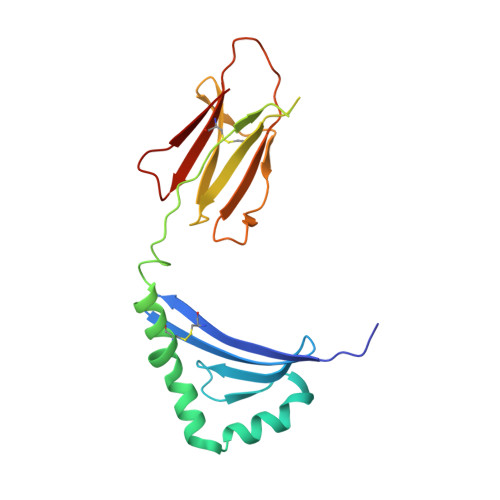
CD4 + T cell-mediated recognition of a conserved cholesterol-dependent cytolysin epitope generates broad antibacterial immunity.
Ciacchi, L., van de Garde, M.D.B., Ladell, K., Farenc, C., Poelen, M.C.M., Miners, K.L., Llerena, C., Reid, H.H., Petersen, J., Price, D.A., Rossjohn, J., van Els, C.A.C.M.(2023) Immunity 56: 1082-1097.e6
- PubMed: 37100059
- DOI: https://doi.org/10.1016/j.immuni.2023.03.020
- Primary Citation of Related Structures:
7T2A, 7T2B, 7T2C, 7T2D - PubMed Abstract:
CD4 + T cell-mediated immunity against Streptococcus pneumoniae (pneumococcus) can protect against recurrent bacterial colonization and invasive pneumococcal diseases (IPDs). Although such immune responses are common, the pertinent antigens have remained elusive. We identified an immunodominant CD4 + T cell epitope derived from pneumolysin (Ply), a member of the bacterial cholesterol-dependent cytolysins (CDCs). This epitope was broadly immunogenic as a consequence of presentation by the pervasive human leukocyte antigen (HLA) allotypes DPB1 ∗ 02 and DPB1 ∗ 04 and recognition via architecturally diverse T cell receptors (TCRs). Moreover, the immunogenicity of Ply 427-444 was underpinned by core residues in the conserved undecapeptide region (ECTGLAWEWWR), enabling cross-recognition of heterologous bacterial pathogens expressing CDCs. Molecular studies further showed that HLA-DP4-Ply 427-441 was engaged similarly by private and public TCRs. Collectively, these findings reveal the mechanistic determinants of near-global immune focusing on a trans-phyla bacterial epitope, which could inform ancillary strategies to combat various life-threatening infectious diseases, including IPDs.
- Infection and Immunity Program, Department of Biochemistry and Molecular Biology, Monash Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia.
Organizational Affiliation: