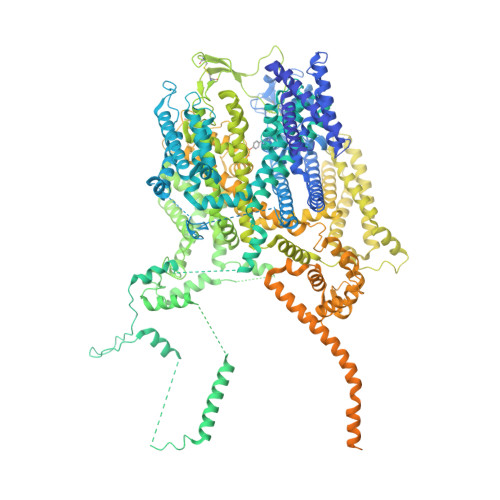
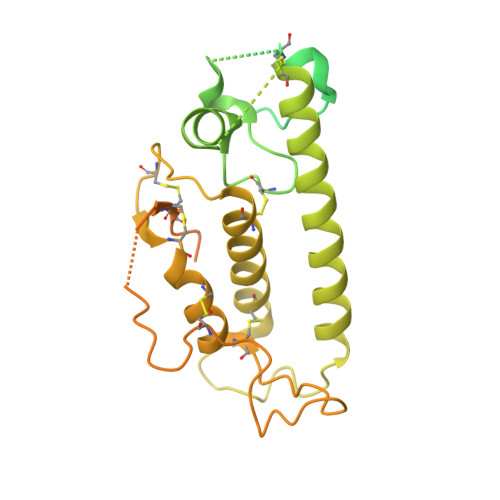
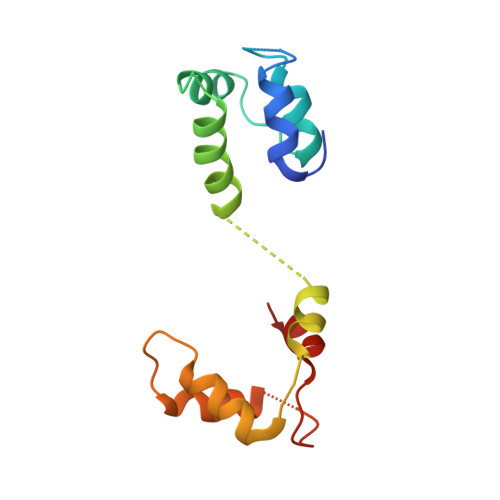
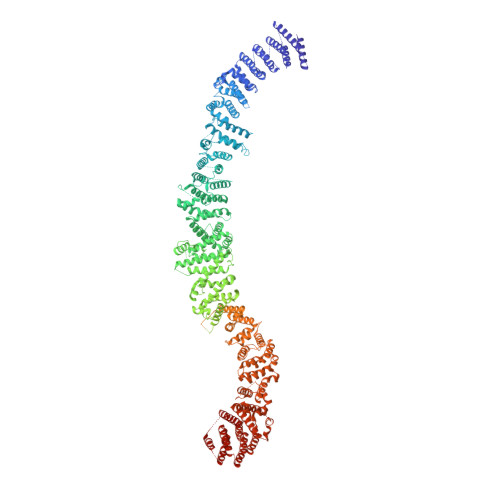
Structural architecture of the human NALCN channelosome.
Kschonsak, M., Chua, H.C., Weidling, C., Chakouri, N., Noland, C.L., Schott, K., Chang, T., Tam, C., Patel, N., Arthur, C.P., Leitner, A., Ben-Johny, M., Ciferri, C., Pless, S.A., Payandeh, J.(2022) Nature 603: 180-186
- PubMed: 34929720
- DOI: https://doi.org/10.1038/s41586-021-04313-5
- Primary Citation of Related Structures:
7SX3, 7SX4 - PubMed Abstract:
Depolarizing sodium (Na + ) leak currents carried by the NALCN channel regulate the resting membrane potential of many neurons to modulate respiration, circadian rhythm, locomotion and pain sensitivity 1-8 . NALCN requires FAM155A, UNC79 and UNC80 to function, but the role of these auxiliary subunits is not understood 3,7,9-12 . NALCN, UNC79 and UNC80 are essential in rodents 2,9,13 , and mutations in human NALCN and UNC80 cause severe developmental and neurological disease 14,15 . Here we determined the structure of the NALCN channelosome, an approximately 1-MDa complex, as fundamental aspects about the composition, assembly and gating of this channelosome remain obscure. UNC79 and UNC80 are massive HEAT-repeat proteins that form an intertwined anti-parallel superhelical assembly, which docks intracellularly onto the NALCN-FAM155A pore-forming subcomplex. Calmodulin copurifies bound to the carboxy-terminal domain of NALCN, identifying this region as a putative modulatory hub. Single-channel analyses uncovered a low open probability for the wild-type complex, highlighting the tightly closed S6 gate in the structure, and providing a basis to interpret the altered gating properties of disease-causing variants. Key constraints between the UNC79-UNC80 subcomplex and the NALCN DI-DII and DII-DIII linkers were identified, leading to a model of channelosome gating. Our results provide a structural blueprint to understand the physiology of the NALCN channelosome and a template for drug discovery to modulate the resting membrane potential.
- Department of Structural Biology, Genentech Inc., South San Francisco, CA, USA.
Organizational Affiliation: