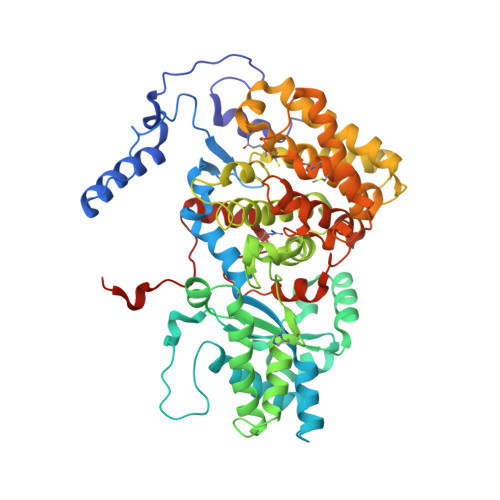
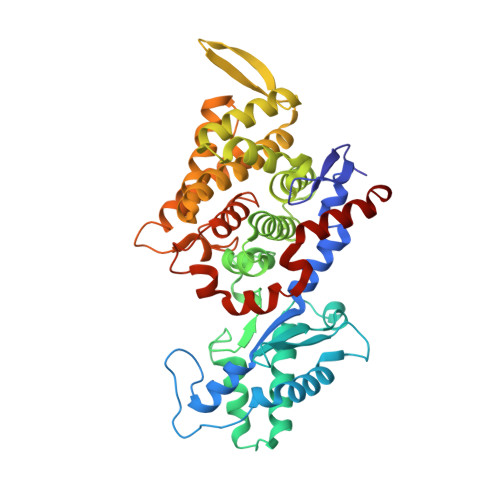
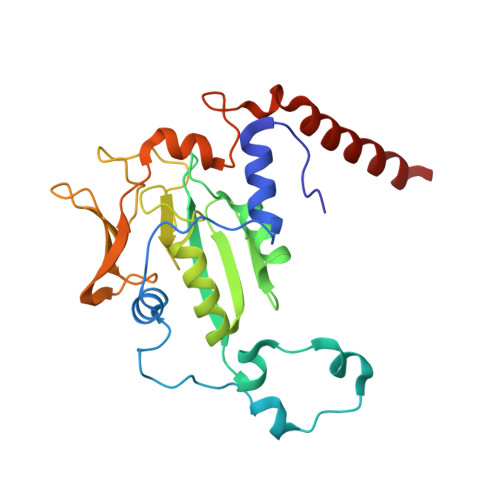
XFEL serial crystallography reveals the room temperature structure of methyl-coenzyme M reductase.
Ohmer, C.J., Dasgupta, M., Patwardhan, A., Bogacz, I., Kaminsky, C., Doyle, M.D., Chen, P.Y., Keable, S.M., Makita, H., Simon, P.S., Massad, R., Fransson, T., Chatterjee, R., Bhowmick, A., Paley, D.W., Moriarty, N.W., Brewster, A.S., Gee, L.B., Alonso-Mori, R., Moss, F., Fuller, F.D., Batyuk, A., Sauter, N.K., Bergmann, U., Drennan, C.L., Yachandra, V.K., Yano, J., Kern, J.F., Ragsdale, S.W.(2022) J Inorg Biochem 230: 111768-111768
- PubMed: 35202981
- DOI: https://doi.org/10.1016/j.jinorgbio.2022.111768
- Primary Citation of Related Structures:
7SUC, 7SXM - PubMed Abstract:
Methyl-Coenzyme M Reductase (MCR) catalyzes the biosynthesis of methane in methanogenic archaea, using a catalytic Ni-centered Cofactor F430 in its active site. It also catalyzes the reverse reaction, that is, the anaerobic activation and oxidation, including the cleavage of the CH bond in methane. Because methanogenesis is the major source of methane on earth, understanding the reaction mechanism of this enzyme can have massive implications in global energy balances. While recent publications have proposed a radical-based catalytic mechanism as well as novel sulfonate-based binding modes of MCR for its native substrates, the structure of the active state of MCR, as well as a complete characterization of the reaction, remain elusive. Previous attempts to structurally characterize the active MCR-Ni(I) state have been unsuccessful due to oxidation of the redox- sensitive catalytic Ni center. Further, while many cryo structures of the inactive Ni(II)-enzyme in various substrates-bound forms have been published, no room temperature structures have been reported, and the structure and mechanism of MCR under physiologically relevant conditions is not known. In this study, we report the first room temperature structure of the MCRred1-silent Ni(II) form using an X-ray Free-Electron Laser (XFEL), with simultaneous X-ray Emission Spectroscopy (XES) and X-ray Diffraction (XRD) data collection. In celebration of the seminal contributions of inorganic chemist Dick Holm to our understanding of nickel-based catalysis, we are honored to announce our findings in this special issue dedicated to this remarkable pioneer of bioinorganic chemistry.
- Department of Biological Chemistry, University of Michigan Medical School, 1150 W. Medical Center Dr., 5200 MSRBIII, Ann Arbor, MI 48109-0606, USA.
Organizational Affiliation: