The crystal structure of CbpD clarifies substrate-specificity motifs in chitin-active lytic polysaccharide monooxygenases.
Dade, C.M., Douzi, B., Cambillau, C., Ball, G., Voulhoux, R., Forest, K.T.(2022) Acta Crystallogr D Struct Biol 78: 1064-1078
- PubMed: 35916229
- DOI: https://doi.org/10.1107/S2059798322007033
- Primary Citation of Related Structures:
7SQX - PubMed Abstract:
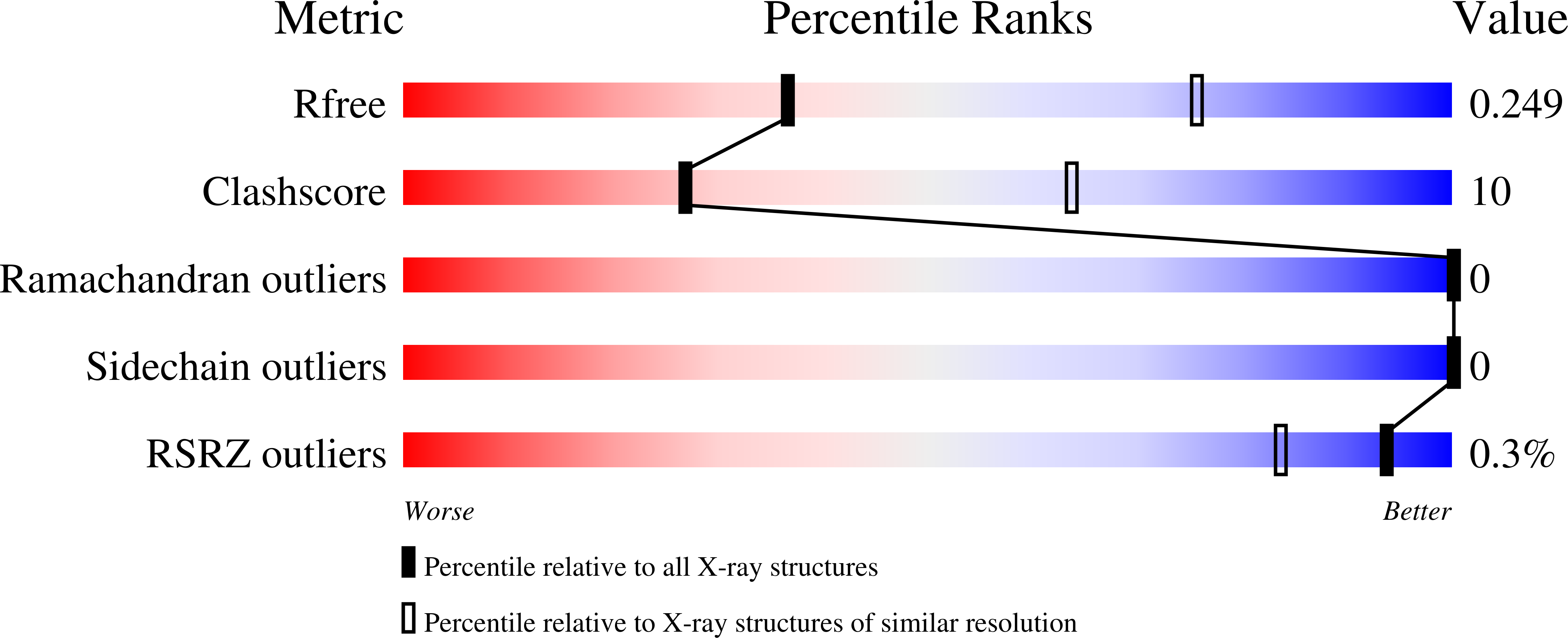
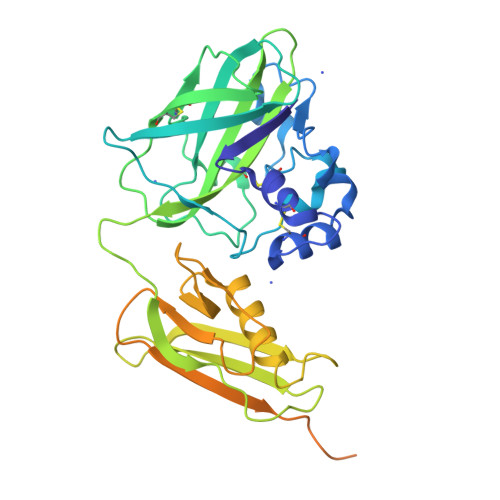
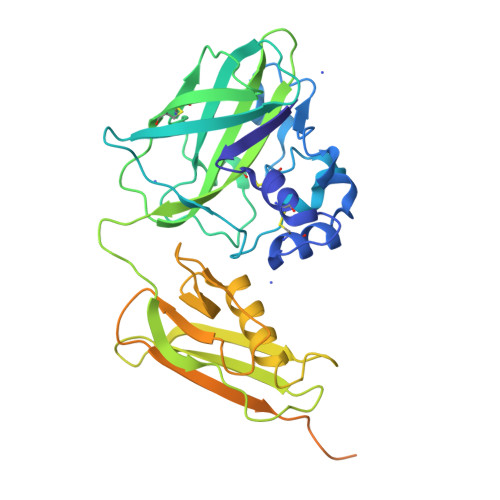
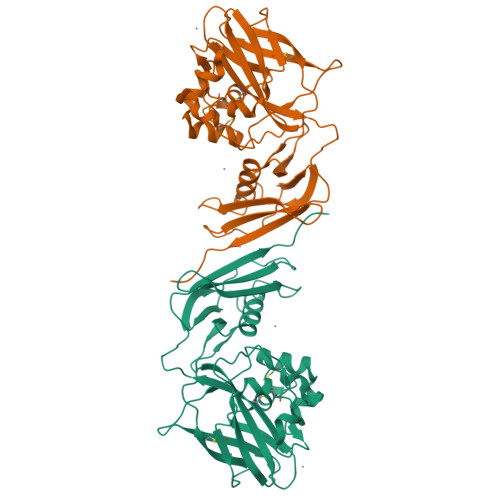
Pseudomonas aeruginosa secretes diverse proteins via its type 2 secretion system, including a 39 kDa chitin-binding protein, CbpD. CbpD has recently been shown to be a lytic polysaccharide monooxygenase active on chitin and to contribute substantially to virulence. To date, no structure of this virulence factor has been reported. Its first two domains are homologous to those found in the crystal structure of Vibrio cholerae GbpA, while the third domain is homologous to the NMR structure of the CBM73 domain of Cellvibrio japonicus CjLPMO10A. Here, the 3.0 Å resolution crystal structure of CbpD solved by molecular replacement is reported, which required ab initio models of each CbpD domain generated by the artificial intelligence deep-learning structure-prediction algorithm RoseTTAFold. The structure of CbpD confirms some previously reported substrate-specificity motifs among LPMOAA10s, while challenging the predictive power of others. Additionally, the structure of CbpD shows that post-translational modifications occur on the chitin-binding surface. Moreover, the structure raises interesting possibilities about how type 2 secretion-system substrates may interact with the secretion machinery and demonstrates the utility of new artificial intelligence protein structure-prediction algorithms in making challenging structural targets tractable.
Organizational Affiliation:
Department of Chemistry, University of Wisconsin-Madison, Madison, Wisconsin, USA.