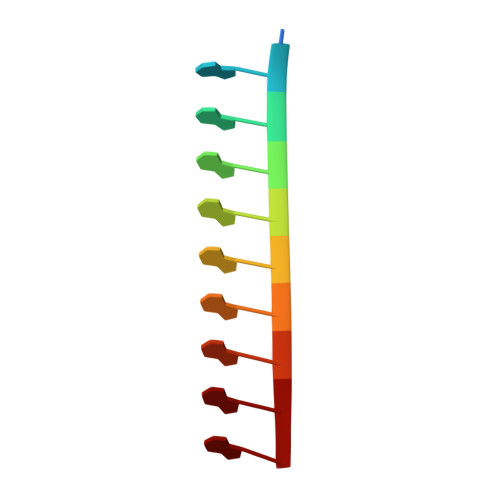
Cryo-EM structure of RNA-induced tau fibrils reveals a small C-terminal core that may nucleate fibril formation.
Abskharon, R., Sawaya, M.R., Boyer, D.R., Cao, Q., Nguyen, B.A., Cascio, D., Eisenberg, D.S.(2022) Proc Natl Acad Sci U S A 119: e2119952119-e2119952119
- PubMed: 35377792
- DOI: https://doi.org/10.1073/pnas.2119952119
- Primary Citation of Related Structures:
7SP1 - PubMed Abstract:
In neurodegenerative diseases including Alzheimer’s and amyotrophic lateral sclerosis, proteins that bind RNA are found in aggregated forms in autopsied brains. Evidence suggests that RNA aids nucleation of these pathological aggregates; however, the mechanism has not been investigated at the level of atomic structure. Here, we present the 3.4-Å resolution structure of fibrils of full-length recombinant tau protein in the presence of RNA, determined by electron cryomicroscopy (cryo-EM). The structure reveals the familiar in-register cross-β amyloid scaffold but with a small fibril core spanning residues Glu391 to Ala426, a region disordered in the fuzzy coat in all previously studied tau polymorphs. RNA is bound on the fibril surface to the positively charged residues Arg406 and His407 and runs parallel to the fibril axis. The fibrils dissolve when RNase is added, showing that RNA is necessary for fibril integrity. While this structure cannot exist simultaneously with the tau fibril structures extracted from patients’ brains, it could conceivably account for the nucleating effects of RNA cofactors followed by remodeling as fibrils mature.
- Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095-1570.
Organizational Affiliation: