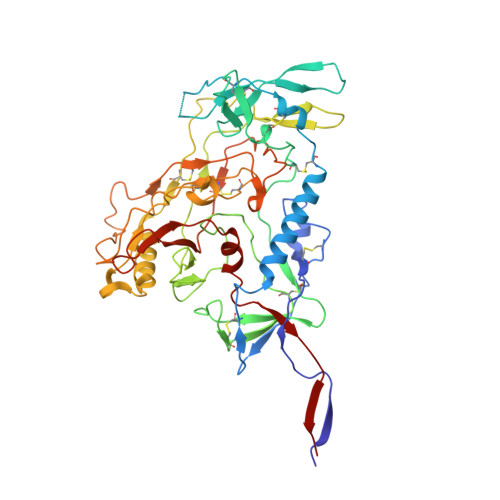
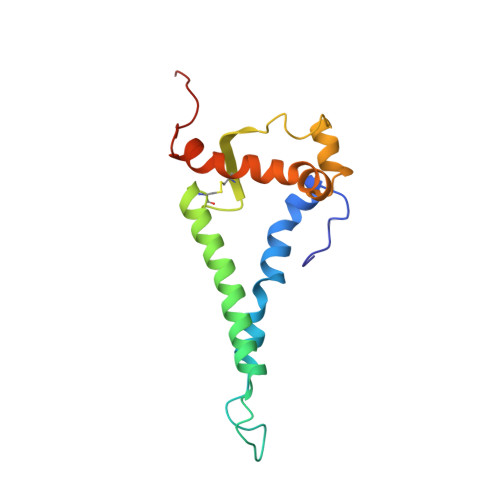
Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice.
Mangala Prasad, V., Leaman, D.P., Lovendahl, K.N., Croft, J.T., Benhaim, M.A., Hodge, E.A., Zwick, M.B., Lee, K.K.(2022) Cell 185: 641-653.e17
- PubMed: 35123651
- DOI: https://doi.org/10.1016/j.cell.2022.01.013
- Primary Citation of Related Structures:
7SKA - PubMed Abstract:
HIV-1 Env mediates viral entry into host cells and is the sole target for neutralizing antibodies. However, Env structure and organization in its native virion context has eluded detailed characterization. Here, we used cryo-electron tomography to analyze Env in mature and immature HIV-1 particles. Immature particles showed distinct Env positioning relative to the underlying Gag lattice, providing insights into long-standing questions about Env incorporation. A 9.1-Å sub-tomogram-averaged reconstruction of virion-bound Env in conjunction with structural mass spectrometry revealed unexpected features, including a variable central core of the gp41 subunit, heterogeneous glycosylation between protomers, and a flexible stalk that allows Env tilting and variable exposure of neutralizing epitopes. Together, our results provide an integrative understanding of HIV assembly and structural variation in Env antigen presentation.
- Department of Medicinal Chemistry, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: