A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection.
Zhou, P., Yuan, M., Song, G., Beutler, N., Shaabani, N., Huang, D., He, W.T., Zhu, X., Callaghan, S., Yong, P., Anzanello, F., Peng, L., Ricketts, J., Parren, M., Garcia, E., Rawlings, S.A., Smith, D.M., Nemazee, D., Teijaro, J.R., Rogers, T.F., Wilson, I.A., Burton, D.R., Andrabi, R.(2022) Sci Transl Med 14: eabi9215-eabi9215
- PubMed: 35133175
- DOI: https://doi.org/10.1126/scitranslmed.abi9215
- Primary Citation of Related Structures:
7SJS - PubMed Abstract:
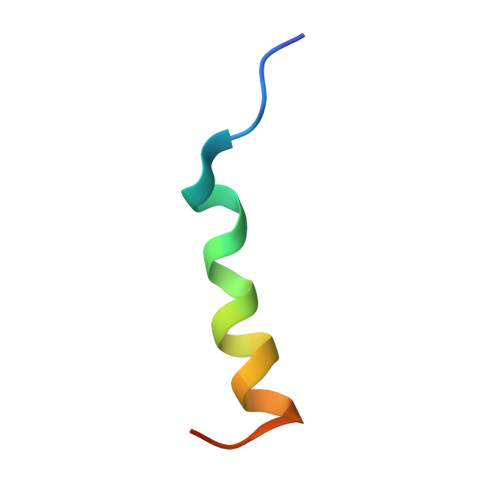
Broadly neutralizing antibodies (bnAbs) to coronaviruses (CoVs) are valuable in their own right as prophylactic and therapeutic reagents to treat diverse CoVs and as templates for rational pan-CoV vaccine design. We recently described a bnAb, CC40.8, from a CoV disease 2019 (COVID-19) convalescent donor that exhibits broad reactivity with human β-CoVs. Here, we showed that CC40.8 targets the conserved S2 stem helix region of the CoV spike fusion machinery. We determined a crystal structure of CC40.8 Fab with a SARS-CoV-2 S2 stem peptide at 1.6-Å resolution and found that the peptide adopted a mainly helical structure. Conserved residues in β-CoVs interacted with CC40.8 antibody, thereby providing a molecular basis for its broad reactivity. CC40.8 exhibited in vivo protective efficacy against SARS-CoV-2 challenge in two animal models. In both models, CC40.8-treated animals exhibited less weight loss and reduced lung viral titers compared to controls. Furthermore, we noted that CC40.8-like bnAbs are relatively rare in human COVID-19 infection, and therefore, their elicitation may require rational structure-based vaccine design strategies. Overall, our study describes a target on β-CoV spike proteins for protective antibodies that may facilitate the development of pan-β-CoV vaccines.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: