Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.
Gade, P., Erlandson, A., Ullah, A., Chen, X., Mathews, I.I., Mera, P.E., Kim, C.Y.(2023) Sci Rep 13: 7980-7980
- PubMed: 37198233
- DOI: https://doi.org/10.1038/s41598-023-34437-9
- Primary Citation of Related Structures:
7SH1 - PubMed Abstract:
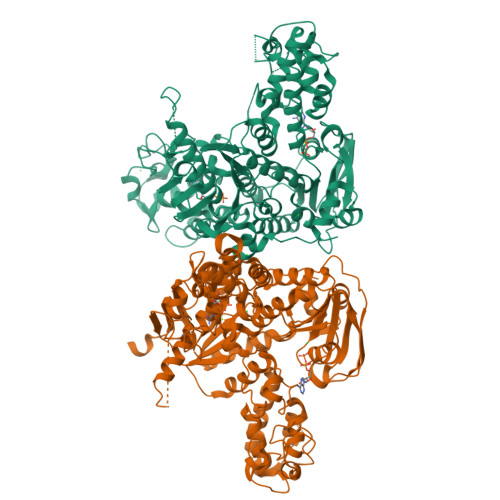
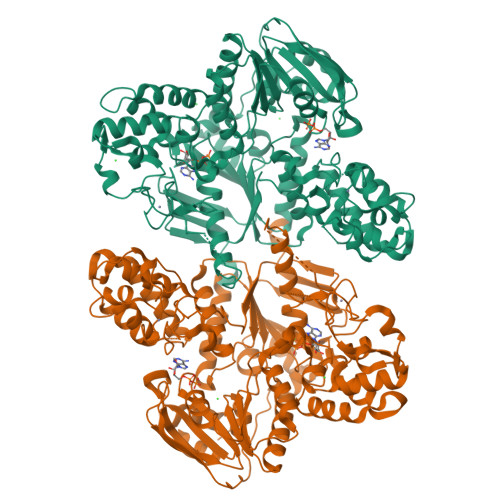
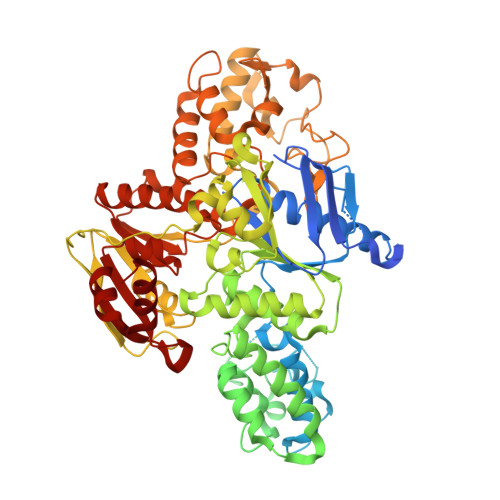
Echinomycin is a natural product DNA bisintercalator antibiotic. The echinomycin biosynthetic gene cluster in Streptomyces lasalocidi includes a gene encoding the self-resistance protein Ecm16. Here, we present the 2.0 Å resolution crystal structure of Ecm16 bound to adenosine diphosphate. The structure of Ecm16 closely resembles that of UvrA, the DNA damage sensor component of the prokaryotic nucleotide excision repair system, but Ecm16 lacks the UvrB-binding domain and its associated zinc-binding module found in UvrA. Mutagenesis study revealed that the insertion domain of Ecm16 is required for DNA binding. Furthermore, the specific amino acid sequence of the insertion domain allows Ecm16 to distinguish echinomycin-bound DNA from normal DNA and link substrate binding to ATP hydrolysis activity. Expression of ecm16 in the heterologous host Brevibacillus choshinensis conferred resistance against echinomycin and other quinomycin antibiotics, including thiocoraline, quinaldopeptin, and sandramycin. Our study provides new insight into how the producers of DNA bisintercalator antibiotics fend off the toxic compounds that they produce.
Organizational Affiliation:
Department of Chemistry and Biochemistry, The University of Texas at El Paso, El Paso, TX, USA.