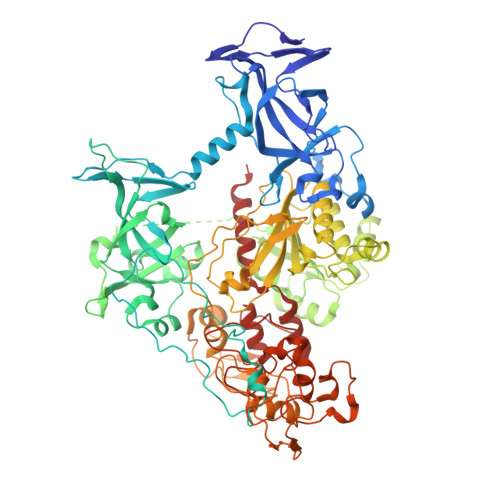
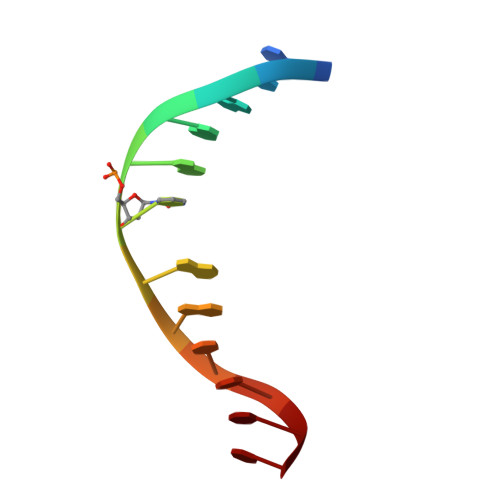
Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Horton, J.R., Pathuri, S., Wong, K., Ren, R., Rueda, L., Fosbenner, D.T., Heerding, D.A., McCabe, M.T., Pappalardi, M.B., Zhang, X., King, B.W., Cheng, X.(2022) Structure 30: 793-802.e5
- PubMed: 35395178
- DOI: https://doi.org/10.1016/j.str.2022.03.009
- Primary Citation of Related Structures:
7SFC, 7SFD, 7SFE, 7SFF, 7SFG - PubMed Abstract:
DNMT1 maintains the parental DNA methylation pattern on newly replicated hemimethylated DNA. The failure of this maintenance process causes aberrant DNA methylation that affects transcription and contributes to the development and progression of cancers such as acute myeloid leukemia. Here, we structurally characterized a set of newly discovered DNMT1-selective, reversible, non-nucleoside inhibitors that bear a core 3,5-dicyanopyridine moiety, as exemplified by GSK3735967, to better understand their mechanism of inhibition. All of the dicyanopydridine-containing inhibitors examined intercalate into the hemimethylated DNA between two CpG base pairs through the DNA minor groove, resulting in conformational movement of the DNMT1 active-site loop. In addition, GSK3735967 introduces two new binding sites, where it interacts with and stabilizes the displaced DNMT1 active-site loop and it occupies an open aromatic cage in which trimethylated histone H4 lysine 20 is expected to bind. Our work represents a substantial step in generating potent, selective, and non-nucleoside inhibitors of DNMT1.
- Department of Epigenetics and Molecular Carcinogenesis, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Organizational Affiliation: