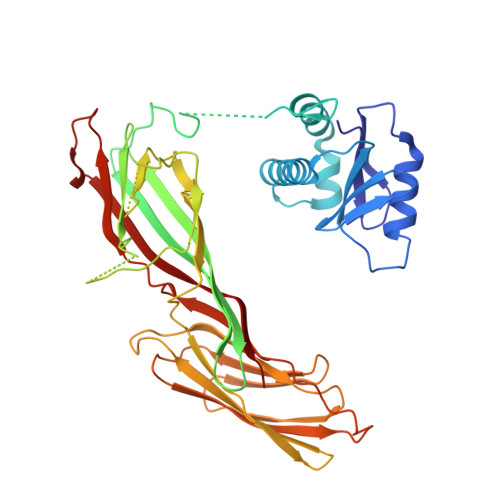
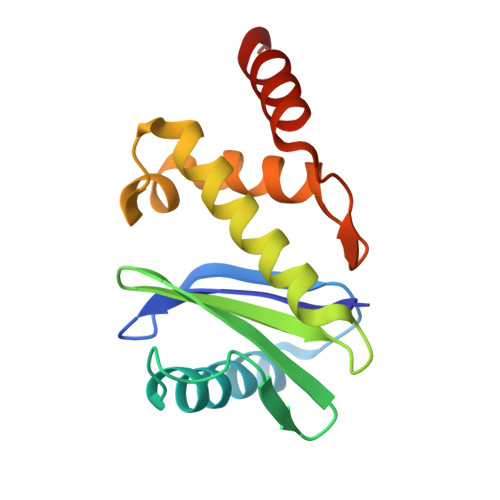
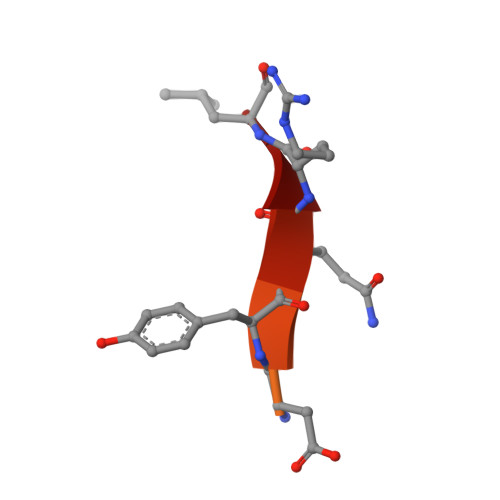
Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Partlow, E.A., Cannon, K.S., Hollopeter, G., Baker, R.W.(2022) Nat Struct Mol Biol 29: 339-347
- PubMed: 35347313
- DOI: https://doi.org/10.1038/s41594-022-00749-z
- Primary Citation of Related Structures:
7RW8, 7RW9, 7RWA, 7RWB, 7RWC - PubMed Abstract:
Clathrin-mediated endocytosis (CME) is the main route of internalization from the plasma membrane. It is known that the heterotetrameric AP2 clathrin adaptor must open to simultaneously engage membrane and endocytic cargo, yet it is unclear how transmembrane cargos are captured to catalyze CME. Using cryogenic-electron microscopy, we discover a new way in which mouse AP2 can reorganize to expose membrane- and cargo-binding pockets, which is not observed in clathrin-coated structures. Instead, it is stimulated by endocytic pioneer proteins called muniscins, which do not enter vesicles. Muniscin-engaged AP2 is primed to rearrange into the vesicle-competent conformation on binding the tyrosine cargo internalization motif (YxxΦ). We propose adaptor priming as a checkpoint to ensure cargo internalization.
- Department of Molecular Medicine, Cornell University, Ithaca, NY, USA.
Organizational Affiliation: