Structural Studies to Enable Drug Discovery
McCue, W.(2021) Thesis
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Caspase-3 subunit p17 | 141 | Homo sapiens | Mutation(s): 0 Gene Names: CASP3, CPP32 | 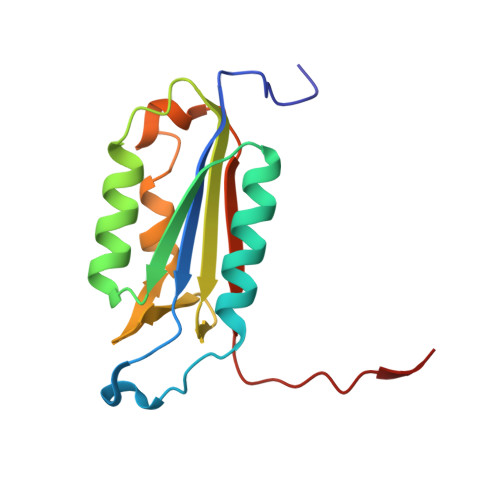 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P42574 (Homo sapiens) Explore P42574 Go to UniProtKB: P42574 | |||||
GTEx: ENSG00000164305 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P42574 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Caspase-3 subunit p12 | 95 | Homo sapiens | Mutation(s): 0 Gene Names: CASP3, CPP32 | 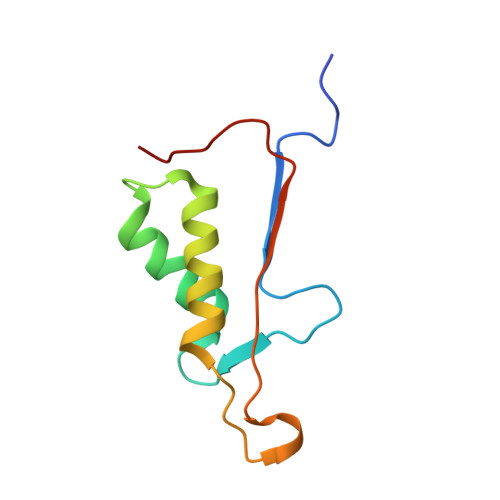 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P42574 (Homo sapiens) Explore P42574 Go to UniProtKB: P42574 | |||||
GTEx: ENSG00000164305 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P42574 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ac-VDVVD-CHO | E [auth F], F [auth G] | 6 | synthetic construct | Mutation(s): 0 | 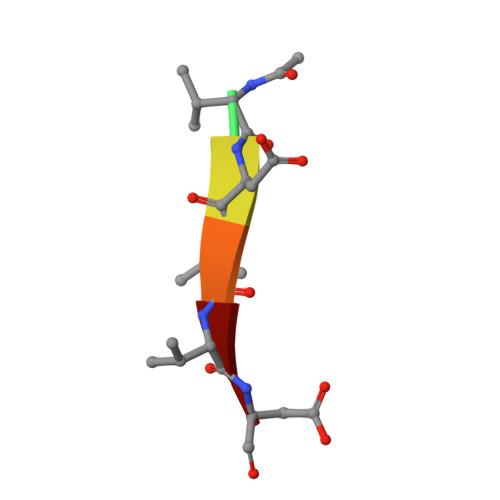 |
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| ASA Query on ASA | E [auth F], F [auth G] | L-PEPTIDE LINKING | C4 H7 N O3 |  | ASP |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.248 | α = 90 |
| b = 85.221 | β = 90 |
| c = 97.621 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| JDirector | data collection |
| PHASER | phasing |
| AutoProcess | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute on Aging (NIH/NIA) | United States | 5R01AG062199-03 |