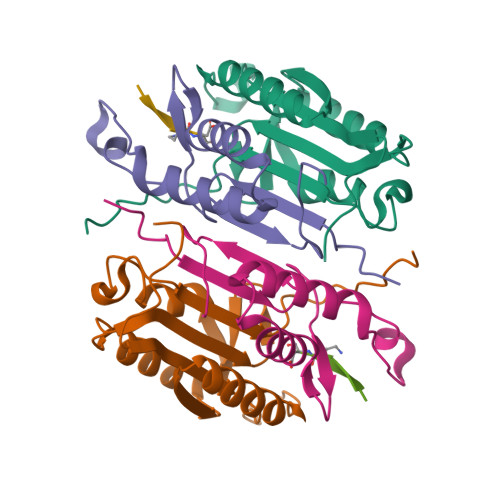
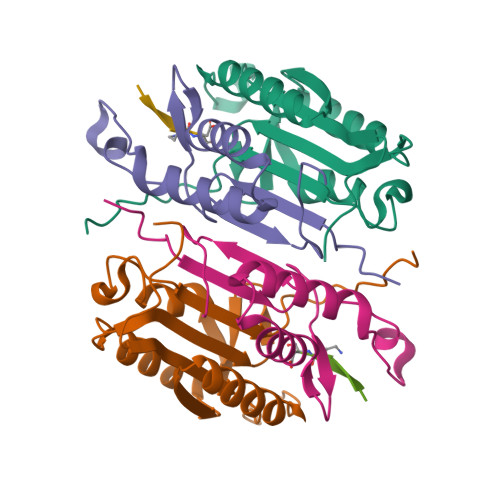
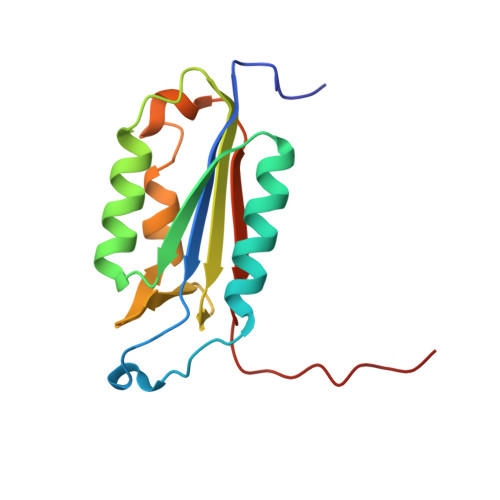
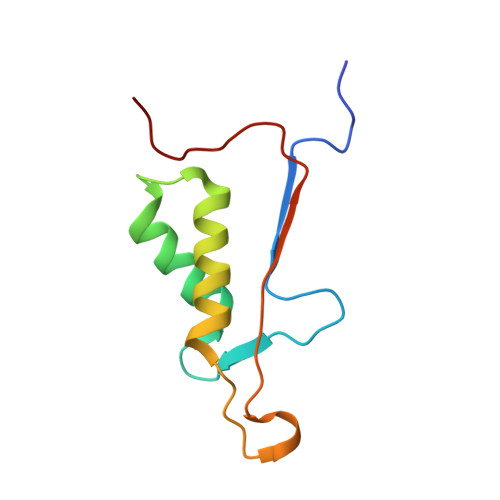
Characterization of caspase-2 inhibitors based on specific sites of caspase-2-mediated proteolysis.
Bresinsky, M., Strasser, J.M., Hubmann, A., Vallaster, B., McCue, W.M., Fuller, J., Singh, G., Nelson, K.M., Cuellar, M.E., Finzel, B.C., Ashe, K.H., Walters, M.A., Pockes, S.(2022) Arch Pharm (Weinheim) 355: e2200095-e2200095
- PubMed: 35642311
- DOI: https://doi.org/10.1002/ardp.202200095
- Primary Citation of Related Structures:
7RNA, 7RNG, 7USO, 7USP, 7USQ - PubMed Abstract:
Since the discovery of the caspase-2 (Casp2)-mediated ∆tau314 cleavage product and its associated impact on tauopathies such as Alzheimer's disease, the design of selective Casp2 inhibitors has become a focus in medicinal chemistry research. In the search for new lead structures with respect to Casp2 selectivity and drug-likeness, we have taken an approach by looking more closely at the specific sites of Casp2-mediated proteolysis. Using seven selected protein cleavage sequences, we synthesized a peptide series of 53 novel molecules and studied them using in vitro pharmacology, molecular modeling, and crystallography. Regarding Casp2 selectivity, AcITV(Dab)D-CHO (23) and AcITV(Dap)D-CHO (26) demonstrated the best selectivity (1-6-fold), although these trends were only moderate. However, some analogous tetrapeptides, most notably AcDKVD-CHO (45), showed significantly increased Casp3 selectivities (>100-fold). Tetra- and tripeptides display decreased or no Casp2 affinity, supporting the assumption that a motif of five amino acids is required for efficient Casp2 inhibition. Overall, the results provide a reasonable basis for the development of both selective Casp2 and Casp3 inhibitors.
Organizational Affiliation:
Institute of Pharmacy, University of Regensburg, Regensburg, Germany.