Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Wagner, S., Fedorov, E., Sudhamalla, B., Jnawali, H.N., Debiec, R., Ghosh, A., Islam, K.(2021) bioRxiv
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2021) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bromodomain-containing protein 3 | 123 | Homo sapiens | Mutation(s): 0 Gene Names: BRD3, KIAA0043, RING3L | 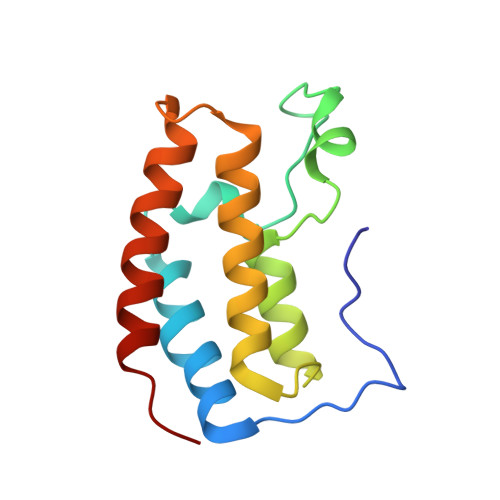 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15059 (Homo sapiens) Explore Q15059 Go to UniProtKB: Q15059 | |||||
PHAROS: Q15059 GTEx: ENSG00000169925 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15059 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine hydroxymethyltransferase, cytosolic | 6 | Homo sapiens | Mutation(s): 0 EC: 2.1.2.1 | 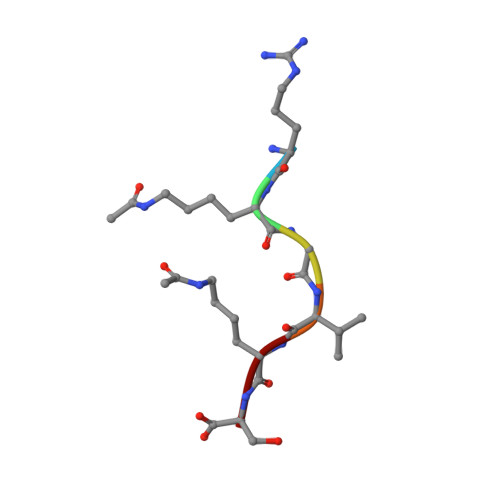 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P34896 (Homo sapiens) Explore P34896 Go to UniProtKB: P34896 | |||||
PHAROS: P34896 GTEx: ENSG00000176974 | |||||
Entity Groups | |||||
| UniProt Group | P34896 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | K [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | E [auth A] F [auth A] G [auth A] H [auth A] I [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| ALY Query on ALY | C, D | L-PEPTIDE LINKING | C8 H16 N2 O3 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 51.55 | α = 90 |
| b = 61.572 | β = 90 |
| c = 83.634 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01GM123234 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01GM130752 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | P01GM118303 |