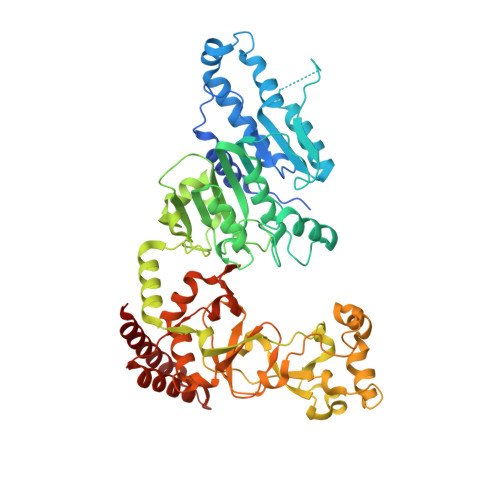
Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Zhou, J., Horton, J.R., Yu, D., Ren, R., Blumenthal, R.M., Zhang, X., Cheng, X.(2022) Epigenetics 17: 970-981
- PubMed: 34523387
- DOI: https://doi.org/10.1080/15592294.2021.1976910
- Primary Citation of Related Structures:
7RFK, 7RFL, 7RFM, 7RFN - PubMed Abstract:
Epigenetically targeted therapeutic development, particularly for SAM-dependent methylations of DNA, mRNA and histones has been proceeding rapidly for cancer treatments over the past few years. However, this approach has barely begun to be exploited for developing new antibiotics, despite an overwhelming global need to counter antimicrobial resistance. Here, we explore whether SAM analogues, some of which are in (pre)clinical studies as inhibitors of human epigenetic enzymes, can also inhibit Clostridioides difficile- specific DNA adenine methyltransferase (CamA), a sporulation regulator present in all C. difficile genomes sequenced to date, but found in almost no other bacteria. We found that SGC0946 (an inhibitor of DOT1L), JNJ-64619178 (an inhibitor of PRMT5) and SGC8158 (an inhibitor of PRMT7) inhibit CamA enzymatic activity in vitro at low micromolar concentrations. Structural investigation of the ternary complexes of CamA-DNA in the presence of SGC0946 or SGC8158 revealed conformational rearrangements of the N-terminal arm, with no apparent disturbance of the active site. This N-terminal arm and its modulation of exchanges between SAM (the methyl donor) and SAH (the reaction product) during catalysis of methyl transfer are, to date, unique to CamA. Our work presents a substantial first step in generating potent and selective inhibitors of CamA that would serve in the near term as chemical probes to investigate the cellular mechanism(s) of CamA in controlling spore formation and colonization, and eventually as therapeutic antivirulence agents useful in treating C. difficile infection.
- Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Organizational Affiliation: