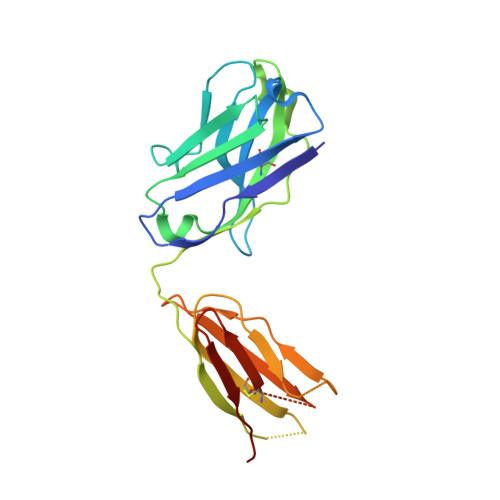
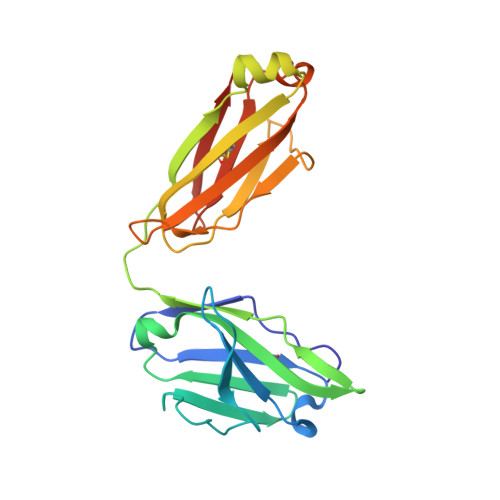
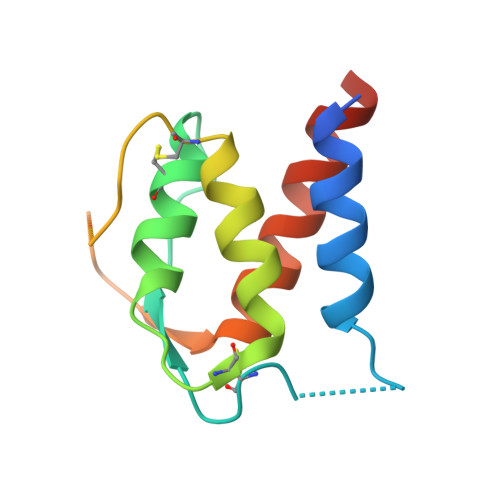
Development of a potent high-affinity human therapeutic antibody via novel application of recombination signal sequence-based affinity maturation.
Kielczewska, A., D'Angelo, I., Amador, M.S., Wang, T., Sudom, A., Min, X., Rathanaswami, P., Pigott, C., Foltz, I.N.(2022) J Biological Chem 298: 101533-101533
- PubMed: 34973336
- DOI: https://doi.org/10.1016/j.jbc.2021.101533
- Primary Citation of Related Structures:
7REW - PubMed Abstract:
Therapeutic antibody development requires discovery of an antibody molecule with desired specificities and drug-like properties. For toxicological studies, a therapeutic antibody must bind the ortholog antigen with a similar affinity to the human target to enable relevant dosing regimens, and antibodies falling short of this affinity design goal may not progress as therapeutic leads. Herein, we report the novel use of mammalian recombination signal sequence (RSS)-directed recombination for complementarity-determining region-targeted protein engineering combined with mammalian display to close the species affinity gap of human interleukin (IL)-13 antibody 731. This fully human antibody has not progressed as a therapeutic in part because of a 400-fold species affinity gap. Using this nonhypothesis-driven affinity maturation method, we generated multiple antibody variants with improved IL-13 affinity, including the highest affinity antibody reported to date (34 fM). Resolution of a cocrystal structure of the optimized antibody with the cynomolgus monkey (or nonhuman primate) IL-13 protein revealed that the RSS-derived mutations introduced multiple successive amino-acid substitutions resulting in a de novo formation of a π-π stacking-based protein-protein interaction between the affinity-matured antibody heavy chain and helix C on IL-13, as well as an introduction of an interface-distant residue, which enhanced the light chain-binding affinity to target. These mutations synergized binding of heavy and light chains to the target protein, resulting in a remarkably tight interaction, and providing a proof of concept for a new method of protein engineering, based on synergizing a mammalian display platform with novel RSS-mediated library generation.
- Amgen British Columbia, Therapeutic Discovery, Burnaby, British Columbia, Canada. Electronic address: akielcze@amgen.com.
Organizational Affiliation: