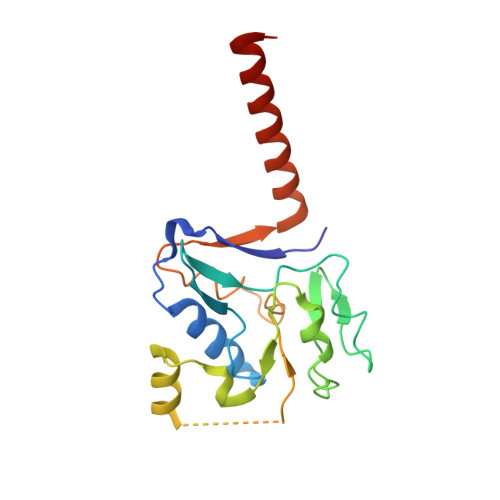
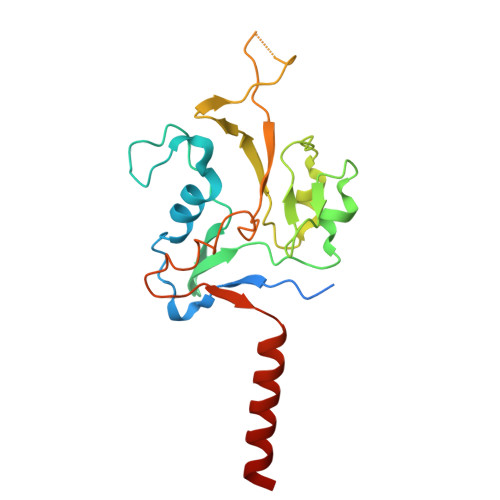
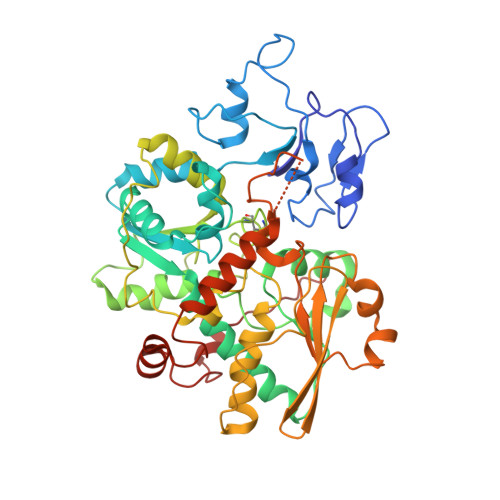
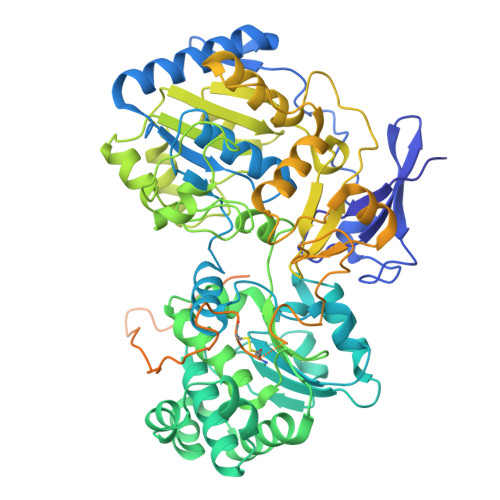
Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Dietrich, H.M., Righetto, R.D., Kumar, A., Wietrzynski, W., Trischler, R., Schuller, S.K., Wagner, J., Schwarz, F.M., Engel, B.D., Muller, V., Schuller, J.M.(2022) Nature 607: 823-830
- PubMed: 35859174
- DOI: https://doi.org/10.1038/s41586-022-04971-z
- Primary Citation of Related Structures:
7QV7 - PubMed Abstract:
Filamentous enzymes have been found in all domains of life, but the advantage of filamentation is often elusive 1 . Some anaerobic, autotrophic bacteria have an unusual filamentous enzyme for CO 2 fixation-hydrogen-dependent CO 2 reductase (HDCR) 2,3 -which directly converts H 2 and CO 2 into formic acid. HDCR reduces CO 2 with a higher activity than any other known biological or chemical catalyst 4,5 , and it has therefore gained considerable interest in two areas of global relevance: hydrogen storage and combating climate change by capturing atmospheric CO 2 . However, the mechanistic basis of the high catalytic turnover rate of HDCR has remained unknown. Here we use cryo-electron microscopy to reveal the structure of a short HDCR filament from the acetogenic bacterium Thermoanaerobacter kivui. The minimum repeating unit is a hexamer that consists of a formate dehydrogenase (FdhF) and two hydrogenases (HydA2) bound around a central core of hydrogenase Fe-S subunits, one HycB3 and two HycB4. These small bacterial polyferredoxin-like proteins oligomerize through their C-terminal helices to form the backbone of the filament. By combining structure-directed mutagenesis with enzymatic analysis, we show that filamentation and rapid electron transfer through the filament enhance the activity of HDCR. To investigate the structure of HDCR in situ, we imaged T. kivui cells with cryo-electron tomography and found that HDCR filaments bundle into large ring-shaped superstructures attached to the plasma membrane. This supramolecular organization may further enhance the stability and connectivity of HDCR to form a specialized metabolic subcompartment within the cell.
- Molecular Microbiology & Bioenergetics, Institute of Molecular Biosciences, Johann Wolfgang Goethe University, Frankfurt am Main, Germany.
Organizational Affiliation: