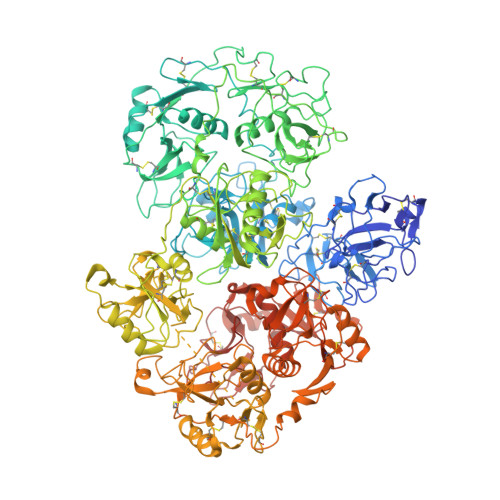
Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch.
Fresquet, M., Lockhart-Cairns, M.P., Rhoden, S.J., Jowitt, T.A., Briggs, D.C., Baldock, C., Brenchley, P.E., Lennon, R.(2022) Proc Natl Acad Sci U S A 119: e2202209119-e2202209119
- PubMed: 35858348
- DOI: https://doi.org/10.1073/pnas.2202209119
- Primary Citation of Related Structures:
7QSR - PubMed Abstract:
Membranous nephropathy is an autoimmune kidney disease caused by autoantibodies targeting antigens present on glomerular podocytes, instigating a cascade leading to glomerular injury. The most prevalent circulating autoantibodies in membranous nephropathy are against phospholipase A2 receptor (PLA2R), a cell surface receptor. The dominant epitope in PLA2R is located within the cysteine-rich domain, yet high-resolution structure-based mapping is lacking. In this study, we define the key nonredundant amino acids in the dominant epitope of PLA2R involved in autoantibody binding. We further describe two essential regions within the dominant epitope and spacer requirements for a synthetic peptide of the epitope for drug discovery. In addition, using cryo-electron microscopy, we have determined the high-resolution structure of PLA2R to 3.4 Å resolution, which shows that the dominant epitope and key residues within the cysteine-rich domain are accessible at the cell surface. In addition, the structure of PLA2R not only suggests a different orientation of domains but also implicates a unique immunogenic signature in PLA2R responsible for inducing autoantibody formation and recognition.
- Wellcome Centre for Cell-Matrix Research, The University of Manchester, Manchester, M13 9PT, United Kingdom.
Organizational Affiliation: