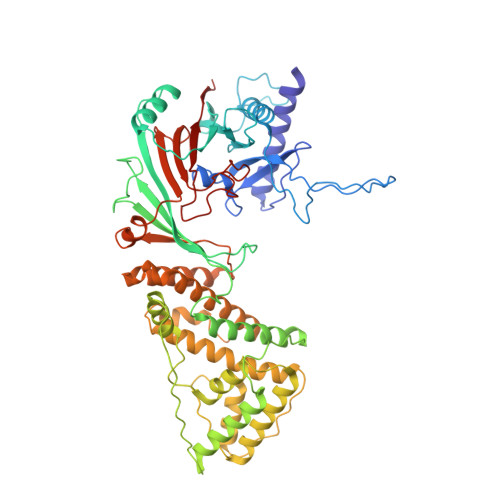
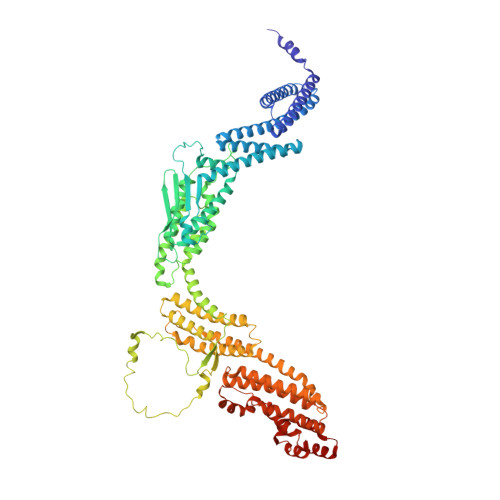
Structure of the RZZ complex and molecular basis of Spindly-driven corona assembly at human kinetochores.
Raisch, T., Ciossani, G., d'Amico, E., Cmentowski, V., Carmignani, S., Maffini, S., Merino, F., Wohlgemuth, S., Vetter, I.R., Raunser, S., Musacchio, A.(2022) EMBO J 41: e110411-e110411
- PubMed: 35373361
- DOI: https://doi.org/10.15252/embj.2021110411
- Primary Citation of Related Structures:
7QPG - PubMed Abstract:
In metazoans, a ≈1 megadalton (MDa) multiprotein complex comprising the dynein-dynactin adaptor Spindly and the ROD-Zwilch-ZW10 (RZZ) complex is the building block of a fibrous biopolymer, the kinetochore fibrous corona. The corona assembles on mitotic kinetochores to promote microtubule capture and spindle assembly checkpoint (SAC) signaling. We report here a high-resolution cryo-EM structure that captures the essential features of the RZZ complex, including a farnesyl-binding site required for Spindly binding. Using a highly predictive in vitro assay, we demonstrate that the SAC kinase MPS1 is necessary and sufficient for corona assembly at supercritical concentrations of the RZZ-Spindly (RZZS) complex, and describe the molecular mechanism of phosphorylation-dependent filament nucleation. We identify several structural requirements for RZZS polymerization in rings and sheets. Finally, we identify determinants of kinetochore localization and corona assembly of Spindly. Our results describe a framework for the long-sought-for molecular basis of corona assembly on metazoan kinetochores.
- Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Dortmund, Germany.
Organizational Affiliation: