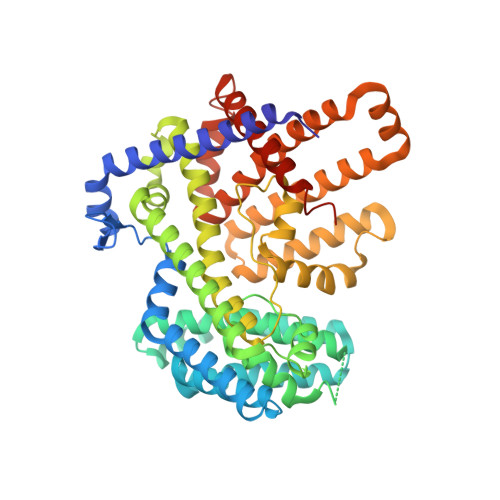
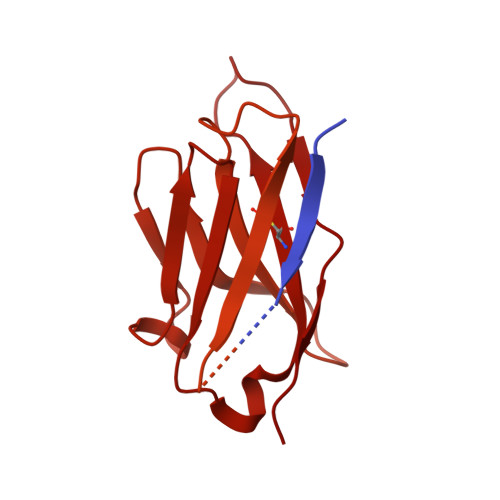
Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Coupland, C.E., Andrei, S.A., Ansell, T.B., Carrique, L., Kumar, P., Sefer, L., Schwab, R.A., Byrne, E.F.X., Pardon, E., Steyaert, J., Magee, A.I., Lanyon-Hogg, T., Sansom, M.S.P., Tate, E.W., Siebold, C.(2021) Mol Cell 81: 5025-5038.e10
- PubMed: 34890564
- DOI: https://doi.org/10.1016/j.molcel.2021.11.018
- Primary Citation of Related Structures:
7Q1U, 7Q6Z - PubMed Abstract:
The Sonic Hedgehog (SHH) morphogen pathway is fundamental for embryonic development and stem cell maintenance and is implicated in various cancers. A key step in signaling is transfer of a palmitate group to the SHH N terminus, catalyzed by the multi-pass transmembrane enzyme Hedgehog acyltransferase (HHAT). We present the high-resolution cryo-EM structure of HHAT bound to substrate analog palmityl-coenzyme A and a SHH-mimetic megabody, revealing a heme group bound to HHAT that is essential for HHAT function. A structure of HHAT bound to potent small-molecule inhibitor IMP-1575 revealed conformational changes in the active site that occlude substrate binding. Our multidisciplinary analysis provides a detailed view of the mechanism by which HHAT adapts the membrane environment to transfer an acyl chain across the endoplasmic reticulum membrane. This structure of a membrane-bound O-acyltransferase (MBOAT) superfamily member provides a blueprint for other protein-substrate MBOATs and a template for future drug discovery.
- Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Organizational Affiliation: