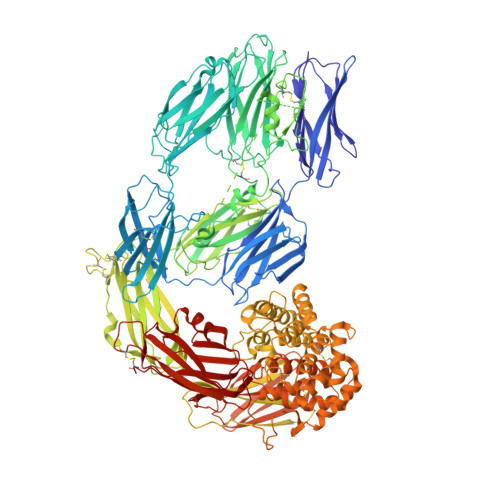
Cryo-EM structures of human A2ML1 elucidate the protease-inhibitory mechanism of the A2M family.
Nielsen, N.S., Zarantonello, A., Harwood, S.L., Jensen, K.T., Kjoge, K., Thogersen, I.B., Schauser, L., Karlsen, J.L., Andersen, G.R., Enghild, J.J.(2022) Nat Commun 13: 3033-3033
- PubMed: 35641520
- DOI: https://doi.org/10.1038/s41467-022-30758-x
- Primary Citation of Related Structures:
7Q1Y, 7Q5Z, 7Q60, 7Q61, 7Q62 - PubMed Abstract:
A2ML1 is a monomeric protease inhibitor belonging to the A2M superfamily of protease inhibitors and complement factors. Here, we investigate the protease-inhibitory mechanism of human A2ML1 and determine the structures of its native and protease-cleaved conformations. The functional inhibitory unit of A2ML1 is a monomer that depends on covalent binding of the protease (mediated by A2ML1's thioester) to achieve inhibition. In contrast to the A2M tetramer which traps proteases in two internal chambers formed by four subunits, in protease-cleaved monomeric A2ML1 disordered regions surround the trapped protease and may prevent substrate access. In native A2ML1, the bait region is threaded through a hydrophobic channel, suggesting that disruption of this arrangement by bait region cleavage triggers the extensive conformational changes that result in protease inhibition. Structural comparisons with complement C3/C4 suggest that the A2M superfamily of proteins share this mechanism for the triggering of conformational change occurring upon proteolytic activation.
- Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark.
Organizational Affiliation: