Development of a multi-targeted chemotherapeutic approach based on G-quadruplex stabilisation and carbonic anhydrase inhibition.
Nocentini, A., Di Porzio, A., Bonardi, A., Bazzicalupi, C., Petreni, A., Biver, T., Bua, S., Marzano, S., Amato, J., Pagano, B., Iaccarino, N., De Tito, S., Amente, S., Supuran, C.T., Randazzo, A., Gratteri, P.(2024) J Enzyme Inhib Med Chem 39: 2366236-2366236
- PubMed: 38905127
- DOI: https://doi.org/10.1080/14756366.2024.2366236
- Primary Citation of Related Structures:
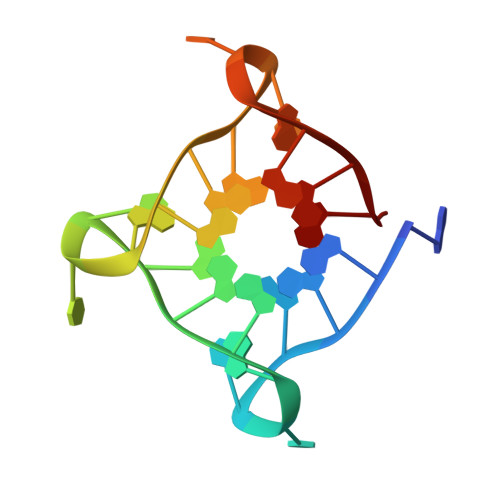
7PNL - PubMed Abstract:
A novel class of compounds designed to hit two anti-tumour targets, G-quadruplex structures and human carbonic anhydrases (hCAs) IX and XII is proposed. The induction/stabilisation of G-quadruplex structures by small molecules has emerged as an anticancer strategy, disrupting telomere maintenance and reducing oncogene expression. hCAs IX and XII are well-established anti-tumour targets, upregulated in many hypoxic tumours and contributing to metastasis. The ligands reported feature a berberine G-quadruplex stabiliser scaffold connected to a moiety inhibiting hCAs IX and XII. In vitro experiments showed that our compounds selectively stabilise G-quadruplex structures and inhibit hCAs IX and XII. The crystal structure of a telomeric G-quadruplex in complex with one of these ligands was obtained, shedding light on the ligand/target interaction mode. The most promising ligands showed significant cytotoxicity against CA IX-positive HeLa cancer cells in hypoxia, and the ability to stabilise G-quadruplexes within tumour cells.
Organizational Affiliation:
NEUROFARBA Department, Pharmaceutical and Nutraceutical Section and Laboratory of Molecular Modeling Cheminformatics & QSAR, University of Florence, Sesto Fiorentino, Florence, Italy.