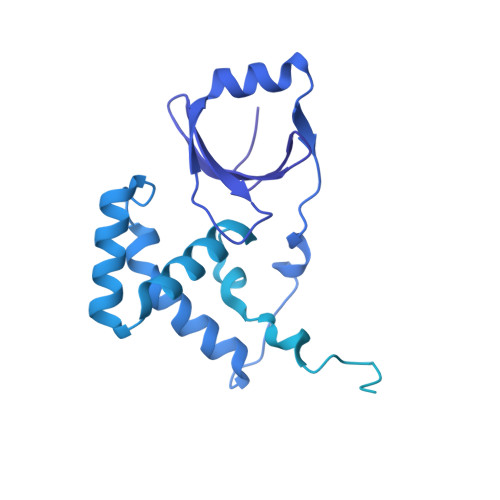
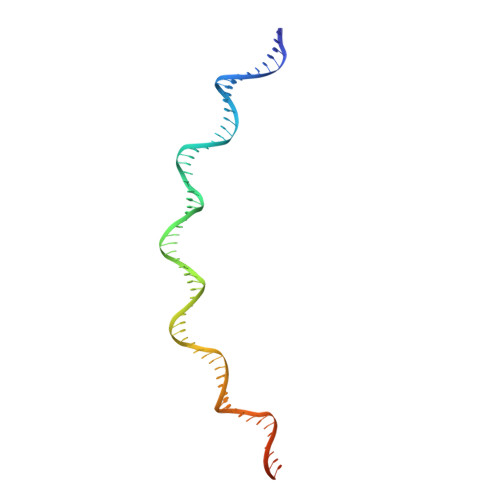
Structural basis of transposon end recognition explains central features of Tn7 transposition systems.
Kaczmarska, Z., Czarnocki-Cieciura, M., Gorecka-Minakowska, K.M., Wingo, R.J., Jackiewicz, J., Zajko, W., Poznanski, J.T., Rawski, M., Grant, T., Peters, J.E., Nowotny, M.(2022) Mol Cell 82: 2618
- PubMed: 35654042
- DOI: https://doi.org/10.1016/j.molcel.2022.05.005
- Primary Citation of Related Structures:
7PIK - PubMed Abstract:
Tn7 is a bacterial transposon with relatives containing element-encoded CRISPR-Cas systems mediating RNA-guided transposon insertion. Here, we present the 2.7 Å cryoelectron microscopy structure of prototypic Tn7 transposase TnsB interacting with the transposon end DNA. When TnsB interacts across repeating binding sites, it adopts a beads-on-a-string architecture, where the DNA-binding and catalytic domains are arranged in a tiled and intertwined fashion. The DNA-binding domains form few base-specific contacts leading to a binding preference that requires multiple weakly conserved sites at the appropriate spacing to achieve DNA sequence specificity. TnsB binding imparts differences in the global structure of the protein-bound DNA ends dictated by the spacing or overlap of binding sites explaining functional differences in the left and right ends of the element. We propose a model of the strand-transfer complex in which the terminal TnsB molecule is rearranged so that its catalytic domain is in a position conducive to transposition.
- Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, 02-109 Warsaw, Poland.
Organizational Affiliation: