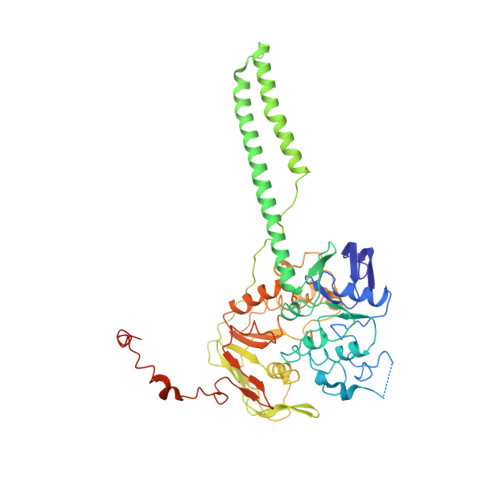
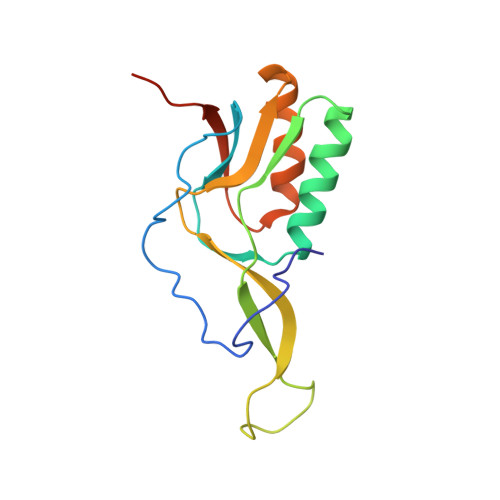
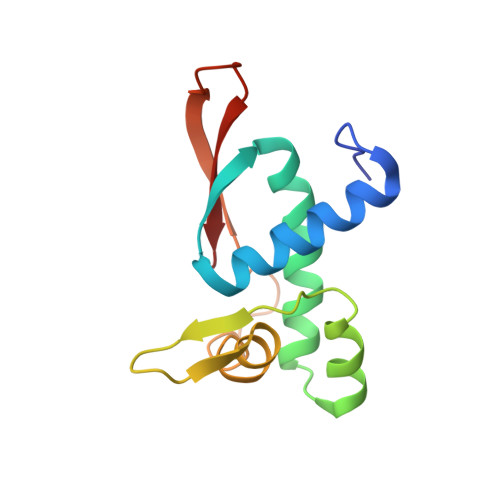
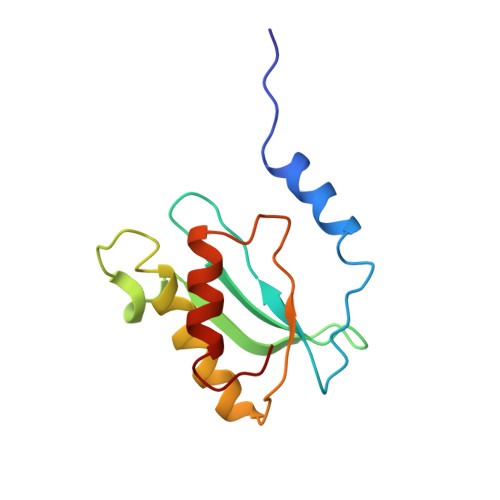
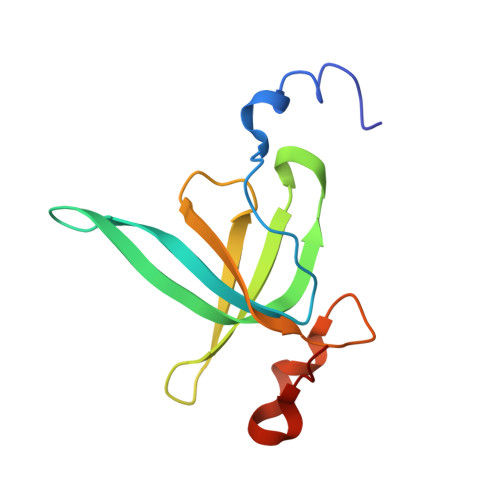
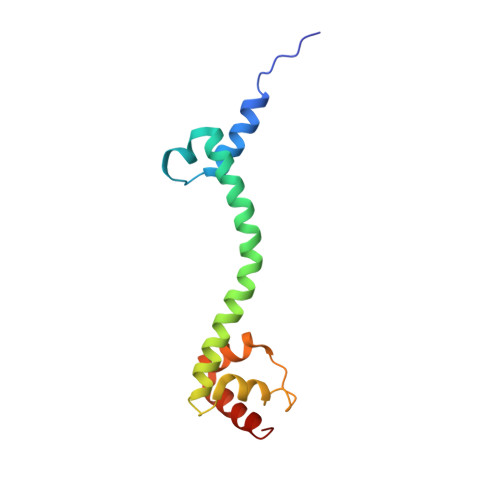
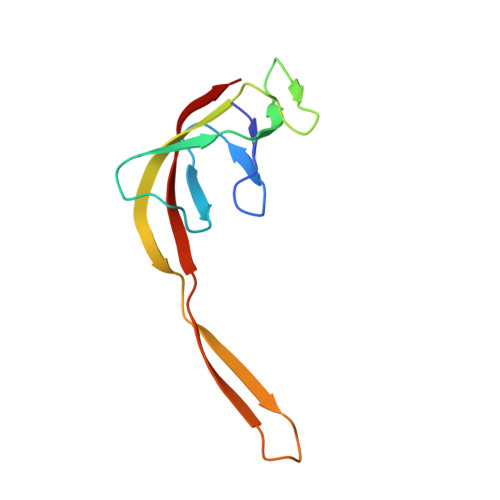
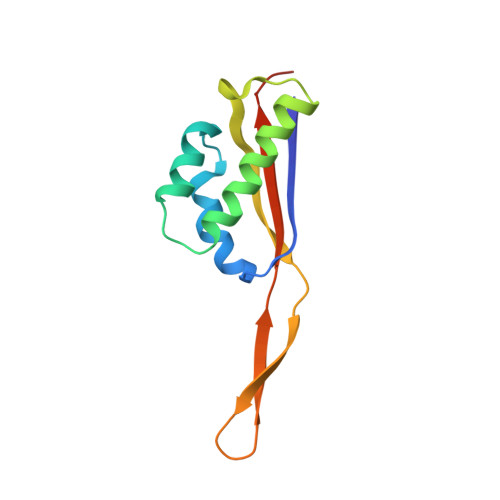
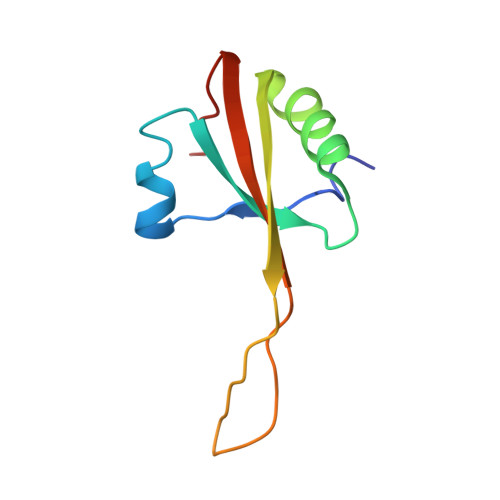
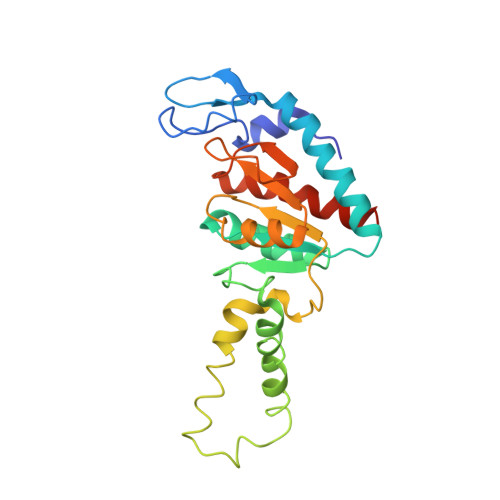
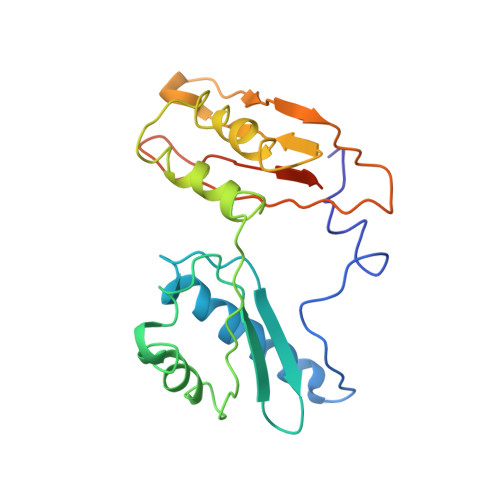
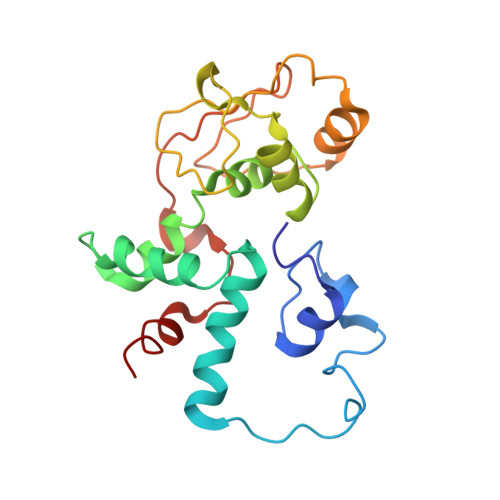
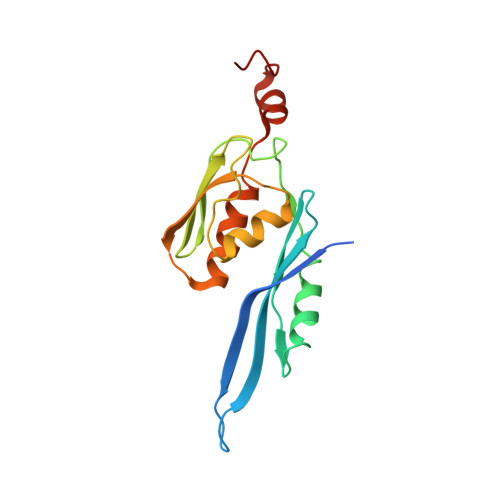
Sal-type ABC-F proteins: intrinsic and common mediators of pleuromutilin resistance by target protection in staphylococci.
Mohamad, M., Nicholson, D., Saha, C.K., Hauryliuk, V., Edwards, T.A., Atkinson, G.C., Ranson, N.A., O'Neill, A.J.(2022) Nucleic Acids Res 50: 2128-2142
- PubMed: 35137182
- DOI: https://doi.org/10.1093/nar/gkac058
- Primary Citation of Related Structures:
7P48 - PubMed Abstract:
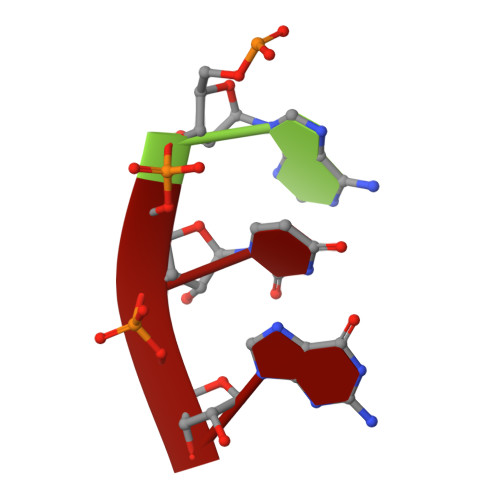
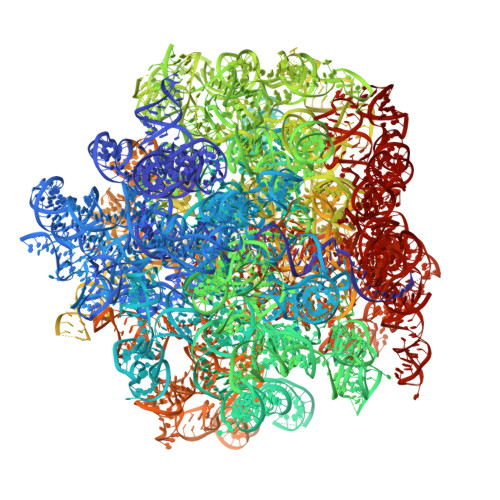
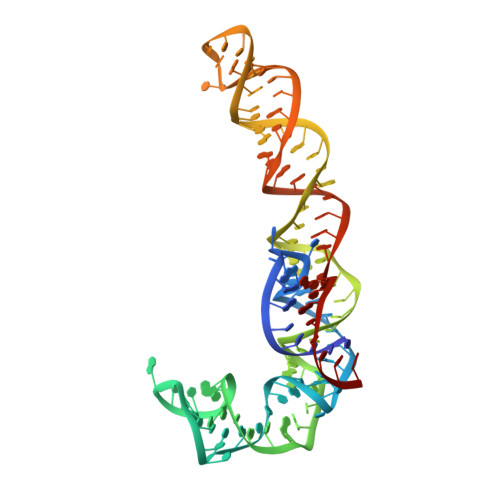
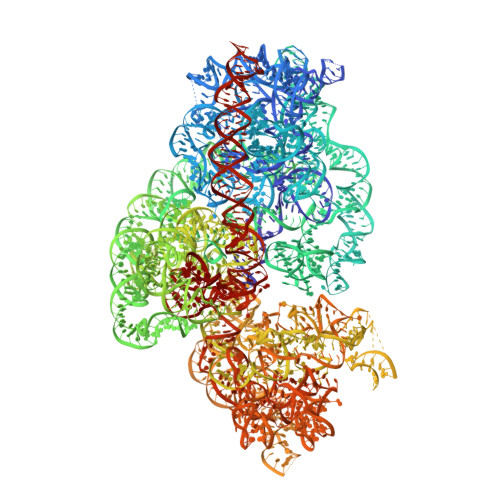
The first member of the pleuromutilin (PLM) class suitable for systemic antibacterial chemotherapy in humans recently entered clinical use, underscoring the need to better understand mechanisms of PLM resistance in disease-causing bacterial genera. Of the proteins reported to mediate PLM resistance in staphylococci, the least-well studied to date is Sal(A), a putative ABC-F NTPase that-by analogy to other proteins of this type-may act to protect the ribosome from PLMs. Here, we establish the importance of Sal proteins as a common source of PLM resistance across multiple species of staphylococci. Sal(A) is revealed as but one member of a larger group of Sal-type ABC-F proteins that vary considerably in their ability to mediate resistance to PLMs and other antibiotics. We find that specific sal genes are intrinsic to particular staphylococcal species, and show that this gene family is likely ancestral to the genus Staphylococcus. Finally, we solve the cryo-EM structure of a representative Sal-type protein (Sal(B)) in complex with the staphylococcal 70S ribosome, revealing that Sal-type proteins bind into the E site to mediate target protection, likely by displacing PLMs and other antibiotics via an allosteric mechanism.
- Astbury Centre for Structural Molecular Biology and School of Molecular & Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, UK.
Organizational Affiliation: