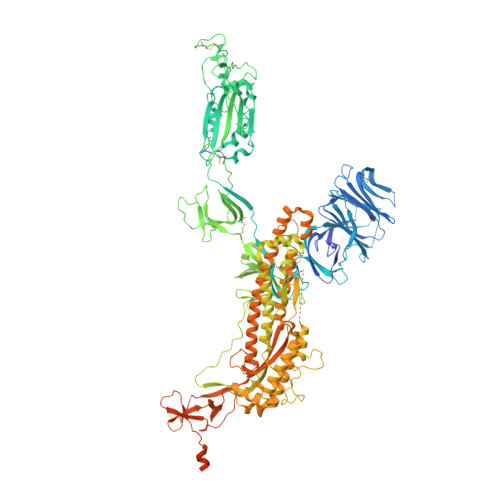
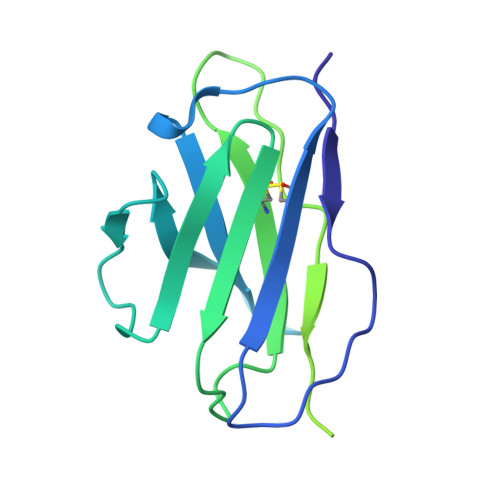
A highly potent antibody effective against SARS-CoV-2 variants of concern.
Fenwick, C., Turelli, P., Perez, L., Pellaton, C., Esteves-Leuenberger, L., Farina, A., Campos, J., Lana, E., Fiscalini, F., Raclot, C., Pojer, F., Lau, K., Demurtas, D., Descatoire, M., Joo, V.S., Foglierini, M., Noto, A., Abdelnabi, R., Foo, C.S., Vangeel, L., Neyts, J., Du, W., Bosch, B.J., Veldman, G., Leyssen, P., Thiel, V., LeGrand, R., Levy, Y., Trono, D., Pantaleo, G.(2021) Cell Rep 37: 109814-109814
- PubMed: 34599871
- DOI: https://doi.org/10.1016/j.celrep.2021.109814
- Primary Citation of Related Structures:
7P40, 7PHG - PubMed Abstract:
Control of the ongoing SARS-CoV-2 pandemic is endangered by the emergence of viral variants with increased transmission efficiency, resistance to marketed therapeutic antibodies, and reduced sensitivity to vaccine-induced immunity. Here, we screen B cells from COVID-19 donors and identify P5C3, a highly potent and broadly neutralizing monoclonal antibody with picomolar neutralizing activity against all SARS-CoV-2 variants of concern (VOCs) identified to date. Structural characterization of P5C3 Fab in complex with the spike demonstrates a neutralizing activity defined by a large buried surface area, highly overlapping with the receptor-binding domain (RBD) surface necessary for ACE2 interaction. We further demonstrate that P5C3 shows complete prophylactic protection in the SARS-CoV-2-infected hamster challenge model. These results indicate that P5C3 opens exciting perspectives either as a prophylactic agent in immunocompromised individuals with poor response to vaccination or as combination therapy in SARS-CoV-2-infected individuals.
- Service of Immunology and Allergy, Department of Medicine, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland.
Organizational Affiliation: