NMR solution structure of SUD-C domain of SARS-CoV-2
Gallo, A., Tsika, A.C., Fourkiotis, N.K., Spyroulias, G.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Non-structural protein 3 | 66 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 3.4.19.12 (PDB Primary Data), 3.4.22 (PDB Primary Data) | 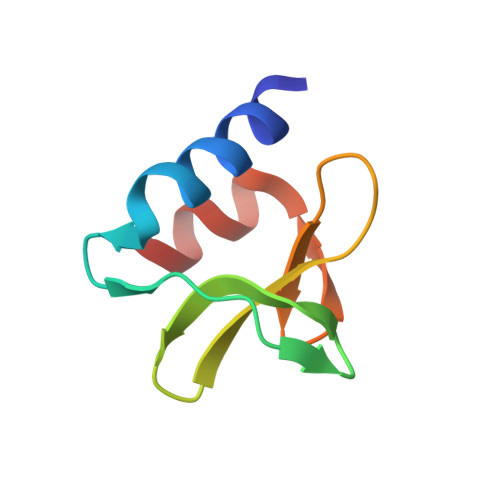 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Regional Development Fund | Greece | 5002550-INSPIRED |
| European Union (EU) | Greece | REGPOT CT-2011-28595-SEEDRUG |