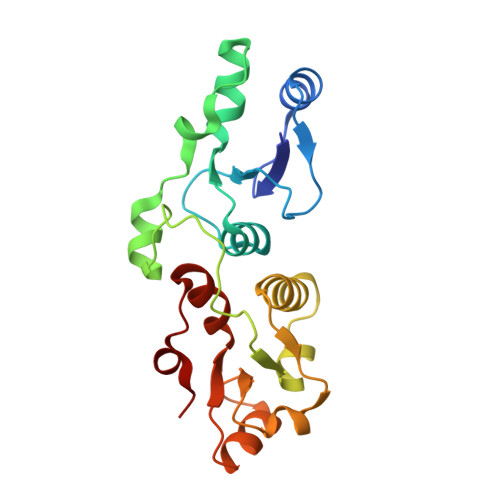
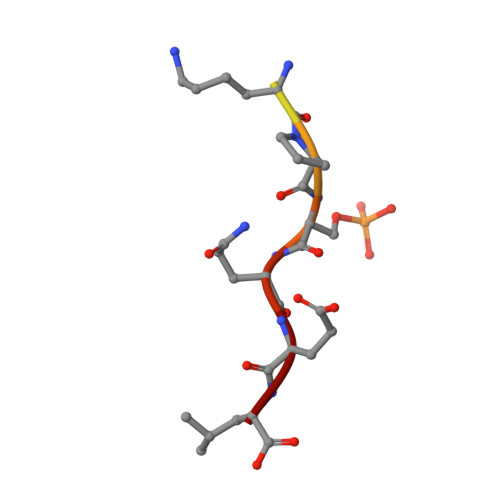
Phosphorylation-dependent assembly of DNA damage response systems and the central roles of TOPBP1.
Day, M., Oliver, A.W., Pearl, L.H.(2021) DNA Repair (Amst) 108: 103232-103232
- PubMed: 34678589
- DOI: https://doi.org/10.1016/j.dnarep.2021.103232
- Primary Citation of Related Structures:
7P0J, 7P0L - PubMed Abstract:
The cellular response to DNA damage (DDR) that causes replication collapse and/or DNA double strand breaks, is characterised by a massive change in the post-translational modifications (PTM) of hundreds of proteins involved in the detection and repair of DNA damage, and the communication of the state of damage to the cellular systems that regulate replication and cell division. A substantial proportion of these PTMs involve targeted phosphorylation, which among other effects, promotes the formation of multiprotein complexes through the specific binding of phosphorylated motifs on one protein, by specialised domains on other proteins. Understanding the nature of these phosphorylation mediated interactions allows definition of the pathways and networks that coordinate the DDR, and helps identify new targets for therapeutic intervention that may be of benefit in the treatment of cancer, where DDR plays a key role. In this review we summarise the present understanding of how phosphorylated motifs are recognised by BRCT domains, which occur in many DDR proteins. We particularly focus on TOPBP1 - a multi-BRCT domain scaffold protein with essential roles in replication and the repair and signalling of DNA damage.
- Cancer Research UK DNA Repair Enzymes Group, Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Falmer, Brighton BN1 9RQ, UK.
Organizational Affiliation: