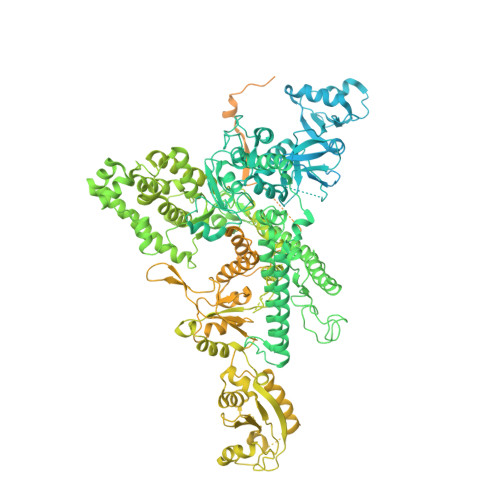
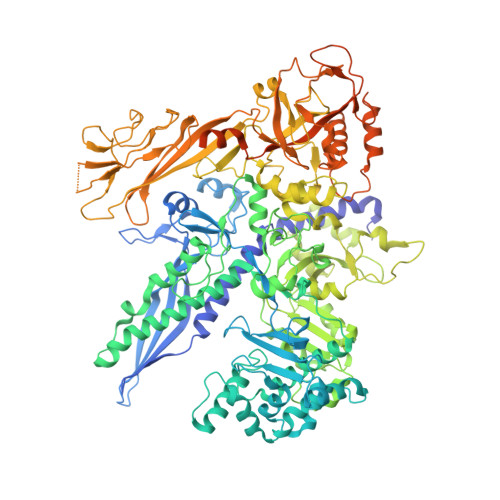
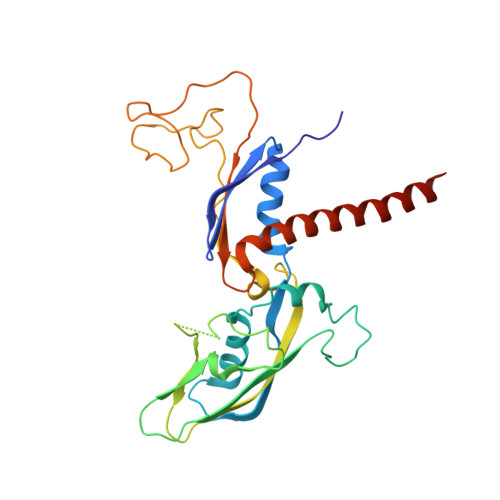
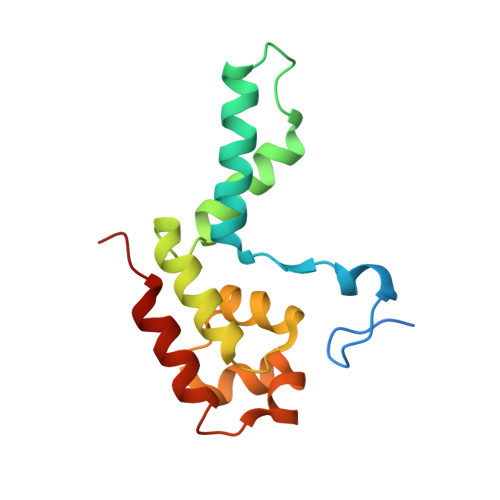
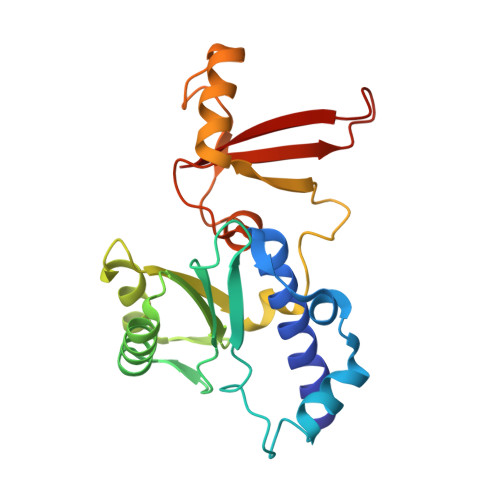
Structure of an inactive RNA polymerase II dimer.
Aibara, S., Dienemann, C., Cramer, P.(2021) Nucleic Acids Res 49: 10747-10755
- PubMed: 34530439
- DOI: https://doi.org/10.1093/nar/gkab783
- Primary Citation of Related Structures:
7OZN, 7OZO, 7OZP - PubMed Abstract:
Eukaryotic gene transcription is carried out by three RNA polymerases: Pol I, Pol II and Pol III. Although it has long been known that Pol I can form homodimers, it is unclear whether and how the two other RNA polymerases dimerize. Here we present the cryo-electron microscopy (cryo-EM) structure of a mammalian Pol II dimer at 3.5 Å resolution. The structure differs from the Pol I dimer and reveals that one Pol II copy uses its RPB4-RPB7 stalk to penetrate the active centre cleft of the other copy, and vice versa, giving rise to a molecular handshake. The polymerase clamp domain is displaced and mobile, and the RPB7 oligonucleotide-binding fold mimics the DNA-RNA hybrid that occupies the cleft during active transcription. The Pol II dimer is incompatible with nucleic acid binding as required for transcription and may represent an inactive storage form of the polymerase.
- Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077, Göttingen, Germany.
Organizational Affiliation: