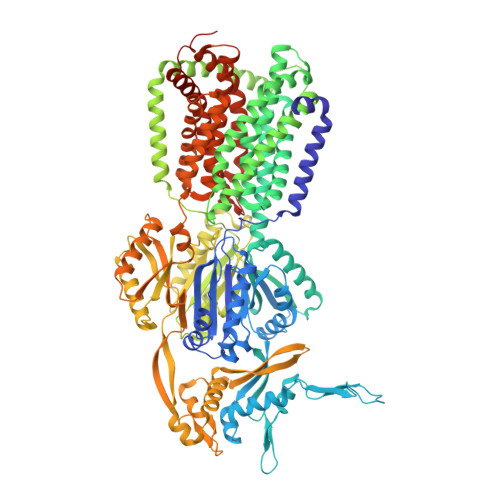
Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps.
Ple, C., Tam, H.K., Vieira Da Cruz, A., Compagne, N., Jimenez-Castellanos, J.C., Muller, R.T., Pradel, E., Foong, W.E., Malloci, G., Ballee, A., Kirchner, M.A., Moshfegh, P., Herledan, A., Herrmann, A., Deprez, B., Willand, N., Vargiu, A.V., Pos, K.M., Flipo, M., Hartkoorn, R.C.(2022) Nat Commun 13: 115-115
- PubMed: 35013254
- DOI: https://doi.org/10.1038/s41467-021-27726-2
- Primary Citation of Related Structures:
7OUK, 7OUL, 7OUM - PubMed Abstract:
Efflux transporters of the RND family confer resistance to multiple antibiotics in Gram-negative bacteria. Here, we identify and chemically optimize pyridylpiperazine-based compounds that potentiate antibiotic activity in E. coli through inhibition of its primary RND transporter, AcrAB-TolC. Characterisation of resistant E. coli mutants and structural biology analyses indicate that the compounds bind to a unique site on the transmembrane domain of the AcrB L protomer, lined by key catalytic residues involved in proton relay. Molecular dynamics simulations suggest that the inhibitors access this binding pocket from the cytoplasm via a channel exclusively present in the AcrB L protomer. Thus, our work unveils a class of allosteric efflux-pump inhibitors that likely act by preventing the functional catalytic cycle of the RND pump.
- Univ. Lille, CNRS, Inserm, CHU Lille, Institut Pasteur Lille, U1019-UMR 9017-CIIL-Center for Infection and Immunity of Lille, F-59000, Lille, France.
Organizational Affiliation: