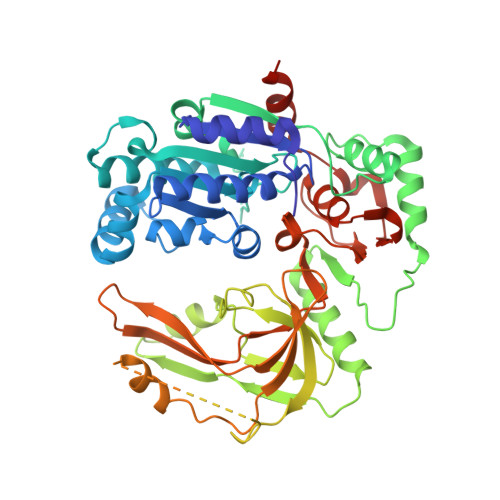
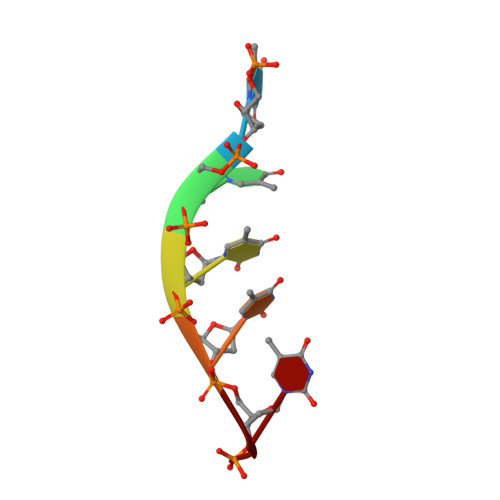
Structural study of the function of Candida Albicans Pif1.
Lu, K.Y., Xin, B.G., Zhang, T., Liu, N.N., Li, D., Rety, S., Xi, X.G.(2021) Biochem Biophys Res Commun 567: 190-194
- PubMed: 34166917
- DOI: https://doi.org/10.1016/j.bbrc.2021.06.050
- Primary Citation of Related Structures:
7OTJ - PubMed Abstract:
Pif1 helicases, conserved in eukaryotes, are involved in maintaining genome stability in both the nucleus and mitochondria. Here, we report the crystal structure of a truncated Candida Albicans Pif1 (CaPif1 368-883 ) in complex with ssDNA and an ATP analog. Our results show that the Q-motif is responsible for identifying adenine bases, and CaPif1 preferentially utilizes ATP/dATP during dsDNA unwinding. Although CaPif1 shares structural similarities with Saccharomyces cerevisiae Pif1, CaPif1 can contact the thymidine bases of DNA by hydrogen bonds, whereas ScPif1 cannot. More importantly, the crosslinking and mutant experiments have demonstrated that the conformational change of domain 2B is necessary for CaPif1 to unwind dsDNA. These findings contribute to further the understanding of the unwinding mechanism of Pif1.
- College of Life Sciences, Northwest A&F University, Yangling, Shaanxi, 712100, China.
Organizational Affiliation: